Abstract
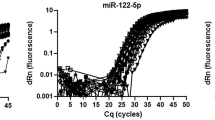
The continuing discoveries of novel classes of RNA modifications in various organisms have raised the need for improving sensitive, convenient, and reliable methods for quantifying RNA modifications. In particular, a subset of small RNAs, including microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs), are modified at their 3′-terminal nucleotides via 2′-O-methylation. However, quantifying the levels of these small RNAs is difficult because 2′-O-methylation at the RNA 3′-terminus inhibits the activity of polyadenylate polymerase and T4 RNA ligase. These two enzymes are indispensable for RNA labeling or ligation in conventional miRNA quantification assays. In this study, we profiled 3′-terminal 2′-O-methyl plant miRNAs in the livers of rice-fed mice by oxidative deep sequencing and detected increasing amounts of plant miRNAs with prolonged oxidation treatment. We further compared the efficiency of stem-loop and poly(A)-tailed RT-qPCR in quantifying plant miRNAs in animal tissues and identified stem-loop RT-qPCR as the only suitable approach. Likewise, stem-loop RT-qPCR was superior to poly(A)-tailed RT-qPCR in quantifying 3′-terminal 2′-O-methyl piRNAs in human seminal plasma. In summary, this study established a standard procedure for quantifying the levels of 3′-terminal 2′-O-methyl miRNAs in plants and piRNAs. Accurate measurement of the 3′-terminal 2′-O-methylation of small RNAs has profound implications for understanding their pathophysiologic roles in biological systems.
Similar content being viewed by others
References
Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res 1994; 22(12): 2183–2196
Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2011; 2(5): 611–631
Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H. MODOMICS: a database of RNA modification pathways 2013 update. Nucleic Acids Res 2013; 41(Database issue D1): D262–D267
Kirino Y, Mourelatos Z. 2′-O-methyl modification in mouse piRNAs and its methylase. Nucleic Acids Symp Ser (Oxf) 2007(51), 417–418
Shen Y, Zheng KX, Duan D, Jiang L, Li J. Label-free microRNA profiling not biased by 3′ end 2′-O-methylation. Anal Chem 2012; 84(15): 6361–6365
Xie Z, Khanna K, Ruan S. Expression of microRNAs and its regulation in plants. Semin Cell Dev Biol 2010; 21(8): 790–797
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116(2): 281–297
Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005; 307(5711): 932–935
Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006; 313(5785): 320–324
Kwon C, Tak H, Rho M, Chang HR, Kim YH, Kim KT, Balch C, Lee EK, Nam S. Detection of PIWI and piRNAs in the mitochondria of mammalian cancer cells. Biochem Biophys Res Commun 2014; 446(1): 218–223
Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011; 12(4): 246–258
Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 2007; 17(14): 1265–1272
Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJT, Roovers EF, Ladurner P, Berezikov E, Ketting RF. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 2010; 29(21): 3688–3700
Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 2007; 13(9): 1397–1401
Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 2007; 21(13): 1603–1608
Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 2009; 25(1): 21–44
Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 2002; 296(5571): 1270–1273
Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 2007; 14(4): 347–348
Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 2009; 15(4): 675–685
Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010; 328(5985): 1534–1539
Aschenbrenner J, Marx A. Direct and site-specific quantification of RNA 2′-O-methylation by PCR with an engineered DNA polymerase. Nucleic Acids Res 2016; 44(8): 3495–3502
Munafó DB, Robb GB. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 2010; 16(12): 2537–2
Hong Y, Wang C, Fu Z, Liang H, Zhang S, Lu M, Sun W, Ye C, Zhang CY, Zen K, Shi L, Zhang C, Chen X. Systematic characterization of seminal plasma piRNAs as molecular biomarkers for male infertility. Sci Rep 2016; 6(1): 24229
Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, Yin Y, Wang C, Zhang T, Zhu D, Zhang D, Xu J, Chen Q, Ba Y, Liu J, Wang Q, Chen J, Wang J, Wang M, Zhang Q, Zhang J, Zen K, Zhang CY. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 2012; 22(1): 107–126
Zhu K, Liu M, Fu Z, Zhou Z, Kong Y, Liang H, Lin Z, Luo J, Zheng H, Wan P, Zhang J, Zen K, Chen J, Hu F, Zhang CY, Ren J, Chen X. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet 2017; 13(8): e1006946
Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu J, Kong H, Zhang Q, Qi X, Hou D, Zhang L, Zhang G, Liu Y, Zhang Y, Li J, Wang J, Chen X, Wang H, Zhang J, Chen H, Zen K, Zhang CY. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res 2015; 25(1): 39–49
Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, Wu X, Wang SE. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res 2016; 26(2): 217–228
Liang G, Zhu Y, Sun B, Shao Y, Jing A, Wang J, Xiao Z. Assessing the survival of exogenous plant microRNA in mice. Food Sci Nutr 2014; 2(4): 380–388
Mlotshwa S, Pruss GJ, MacArthur JL, Endres MW, Davis C, Hofseth LJ, Peña MM, Vance V. A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res 2015; 25(4): 521–524
Yang J, Farmer LM, Agyekum A A A, Elbaz-Younes I, Hirschi KD. Detection of an abundant plant-based small RNA in healthy consumers. PLoS One 2015; 10(9): e0137516
Yang J, Farmer LM, Agyekum A A A, Hirschi KD. Detection of dietary plant-based small RNAs in animals. Cell Res 2015; 25(4): 517–520
Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 2013; 31(11): 965–967
Masood M, Everett CP, Chan SY, Snow JW. Negligible uptake and transfer of diet-derived pollen microRNAs in adult honey bees. RNA Biol 2016; 13(1): 109–118
Tosar JP, Rovira C, Naya H, Cayota A. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA 2014; 20(6): 754–757
Micó V, Martín R, Lasunción MA, Ordovás JM, Daimiel L. Unsuccessful detection of plant microrNAs in beer, extra virgin olive oil and human plasma after an acute ingestion of extra virgin olive oil. Plant Foods Hum Nutr 2016; 71(1): 102–108
Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res 2012; 22(4): 624–636
Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 2007; 14(4): 349–350
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18(10): 997–1006
Haldipur B, Arankalle V. Circulating miR-122 levels in self-recovering hepatitis E patients. ExRNA 2019; 1: 2
Wang N, Qu S, Sun W, Zeng Z, Liang H, Zhang CY, Chen X, Zen K. Direct quantification of 3′ terminal 2′-O-methylation of small RNAs by RT-qPCR. RNA 2018; 24(11): 1520–1529
Zhao C, Sun X, Li L. Biogenesis and function of extracellular miRNAs. ExRNA 2019; 1: 38
Jiang X, Hou D, Wei Z, Zheng S, Zhang Y, Li J. Extracellular and intracellular microRNAs in pancreatic cancer: from early diagnosis to reducing chemoresistance. ExRNA 2019; 1: 17
Perera BPU, Tsai ZT, Colwell ML, Jones TR, Goodrich JM, Wang K, Sartor MA, Faulk C, Dolinoy DC. Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics 2019; 14(5): 504–521
Tang W, Tu S, Lee HC, Weng Z, Mello CC. The ribonuclease PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell 2016; 164(5): 974–984
Sellitto A, Geles K, D’Agostino Y, Conte M, Alexandrova E, Rocco D, Nassa G, Giurato G, Tarallo R, Weisz A, Rizzo F. Molecular and functional characterization of the somatic PIWIL1/piRNA pathway in colorectal cancer cells. Cells 2019; 8(11): 1390
Tang F, Hayashi K, Kaneda M, Lao K, Surani MA. A sensitive multiplex assay for piRNA expression. Biochem Biophys Res Commun 2008; 369(4): 1190–1194
Honda S, Loher P, Morichika K, Shigematsu M, Kawamura T, Kirino Y, Rigoutsos I, Kirino Y. Increasing cell density globally enhances the biogenesis of Piwi-interacting RNAs in Bombyx mori germ cells. Sci Rep 2017; 7(1): 4110
Kang W, Bang-Berthelsen CH, Holm A, Houben AJS, Müller AH, Thymann T, Pociot F, Estivill X, Friedländer MR. Survey of 800+ data sets from human tissue and body fluid reveals xenomiRs are likely artifacts. RNA 2017; 23(4): 433–445
Tóth KF, Pezic D, Stuwe E, Webster A. The piRNA pathway guards the germline genome against transposable elements. Adv Exp Med Biol 2016; 886: 51–77
Zhao K, Cheng S, Miao N, Xu P, Lu X, Zhang Y, Wang M, Ouyang X, Yuan X, Liu W, Lu X, Zhou P, Gu J, Zhang Y, Qiu D, Jin Z, Su C, Peng C, Wang JH, Dong MQ, Wan Y, Ma J, Cheng H, Huang Y, Yu Y. A Pandas complex adapted for piRNA-guided transcriptional silencing and heterochromatin formation. Nat Cell Biol 2019; 21(10): 1261–1272
Sun T, Han X. The disease-related biological functions of PIWI-interacting RNAs (piRNAs) and underlying molecular mechanisms. ExRNA 2019; 1: 21
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (No. 020814380146), National Basic Research Program of China (973 Program) (No. 2014CB542300), National Natural Science Foundation of China (Nos. 32022015, 32001077, 31871295, 21877060, 81250044, 81602697, and 81772727), and Research Unit of Extracellular Non-Coding RNA, Chinese Academy of Medical Sciences (No. 2021RU015).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Compliance with ethics guidelines
Yan Kong, Huanhuan Hu, Yangyang Shan, Zhen Zhou, Ke Zen, Yulu Sun, Rong Yang, Zheng Fu, and Xi Chen declare that they have no conflict of interest. The study protocol was approved by the Institutional Review Board of Nanjing University (Nanjing, China) (Approval No. IACUC-2006008). All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5). Informed consent was obtained from all patients who provided semen samples.
Supplementary files
Rights and permissions
About this article
Cite this article
Kong, Y., Hu, H., Shan, Y. et al. Accurate quantification of 3′-terminal 2′-O-methylated small RNAs by utilizing oxidative deep sequencing and stem-loop RT-qPCR. Front. Med. 16, 240–250 (2022). https://doi.org/10.1007/s11684-021-0909-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11684-021-0909-7