Abstract
Ketosis is a condition where ketone bodies are produced as an alternative energy source, due to insufficient glucose for energy production so that the body switches from carbohydrate metabolism to mostly fat metabolism. In this study, we examined the anti-ketosis effects of silibinin, a major active component of silymarin. We induced ketosis in FL83B mouse hepatocytes in vitro by culturing in low glucose media and compared results to hepatocytes maintained in high-glucose conditions. We quantified β-hydroxybutyrate (BHB) levels with a colorimetric assay. In low-glucose conditions, silibinin reduced the amount of BHB produced, compared to high-glucose conditions; thus, silibinin exhibited an anti-ketotic effect. Ketone body formation during beta oxidation is mediated by 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2). The nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) regulates the transcription of HMGCS2, and plays a vital role in BHB levels. We showed that silibinin inhibited the expression of HMGCS2 and NF-kB at transcriptional and translational levels. Silibinin also inhibited the nuclear translocation of NF-kB and its DNA binding activity. To elucidate the relationship between HMGCS2 and NF-kB, we tested inhibited and over-expressed NF-kB. We found that NF-kB acted as a positive regulator for HMGCS2 under ketosis treatment conditions.
Similar content being viewed by others
Introduction
Induction of in vitro ketosis is a convenient method for testing the efficacy of specific drugs designed to treat ketosis and for determining the molecular targets involved. Liver produces ketone bodies (acetoacetate and β-hydroxybutyrate) from fatty acids in response to fasting, a carbohydrate-restrictive diet, or glucose starvation. The presence of ketone bodies in the bloodstream could cause a relative imbalance in fat metabolism (Garber et al. 1974). The glucose levels in the body are significantly different in normal conditions and ketotic conditions; low-glucose conditions induce the body to use fat as an alternative energy source; hence, ketosis ensues. Thus, the induction of in vitro ketotic conditions is not difficult; it can be achieved by varying the amount of glucose (D-glucose) in the cell culture medium (Sreekantan Preetha et al. 2015).
Ketosis can be treated with natural substances without side effects. In dairy cows, these substances can be mixed into the feed. Natural products that reduce ketone body concentrations to below 0.5 mM (ketotic conditions), with no adverse effects at the required concentrations, are considered suitable anti-ketotic drugs. Silibinin is the major active constituent of silymarin, an extract from milk thistle seeds. Silibinin has shown pharmacological effects, due to its structure, which consists of two diastereomers, silybin A, and silybin B (Davis-Searles et al. 2005). Silibinin showed effective pharmacological activity in the liver, when it was used as a supportive element for alcoholic treatments and when it was used to treat child grade “A” liver cirrhosis (Saller et al. 2008). Silibinin is also an anti-cancer drug; it has shown high efficacy against various cancers, including colon (Kumar et al. 2014), breast (Wang et al. 2010), bladder (Zeng et al. 2011), skin (Singh and Agarwal 2005), and lung (Mateen et al. 2010) cancers.
Ketosis treatment by targeting molecular pathways is the most efficient treatment. Ketone bodies are produced in mitochondria through beta oxidation (Cotter et al. 2014). The mitochondrial enzyme, 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2), converts acetoacetyl-CoA to HMG-CoA, which is then converted to ketone bodies, acetoacetate and β hydroxybutyrate (BHB). In humans, HMGCS2 is encoded by the HMGCS2 gene. In hepatic mitochondria, mitochondrial HMGCS2 produces HMG-CoA, which is then transformed into acetoacetate by HMG-CoA lyase. Finally, acetoacetate is transformed to hydroxybutyrate and acetone (Hegardt 1999). Ketosis can be controlled at the molecular level by inhibiting the expression of HMGSC2, which reduces beta oxidation. Thus, an anti-ketosis drug that inhibits or down-regulates HMGCS2 activity would reduce ketone body production.
Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) is a transcription factor that regulates the expression of cytokines, chemokines, and adhesion molecules (Lawrence 2009). The NF-kB protein complex controls DNA transcription and cell survival. NF-kB is a rapid-acting, primary transcription factor for c-myc (Duyao et al. 1990), CXCL (Tensen et al. 1999), VCAM (Iademarco et al. 1992), CCL2 (Ueda et al. 1994), etc. It also regulates peroxisome proliferator-activated receptor (PPAR) levels by regulating expression of the PGC-1β gene (Abrahan and Ash 2016). NF-kB is a transcription factor for HMGCS2; it can regulate the expression of HMGCS2 by regulating PPAR (Vilà-Brau et al. 2011). However, it remains unknown whether NF-kB acts as a positive or negative regulator of HMGCS2 expression. Understanding this relationship can elucidate a molecular pathway that can be targeted in the treatment of ketosis.
In the present study, we investigated the anti-ketotic activity of silibinin. We analyzed its activity on HMGCS2 and NF-kB, and we explored the possibility of targeting the HMGCS2/NF-kB pathway in the development of treatments for ketosis. We also elucidated how NF-kB acted on HMGCS2 in ketotic conditions.
Materials and Methods
Antibodies and reagents
Colorimetric assay kit for detecting BHB was purchased from the Cayman Chemical Co. (Ann Arbor, MI). Silibinin and D-glucose were purchased from Sigma-Aldrich (St. Louis, MO). F-12K medium and anti-β-actin antibody were purchased from Life Technologies (Carlsbad, CA). Penicillin–streptomycin solution and fetal bovine serum (FBS) were purchased from HyClone (South of Logan, UT). Trypsin-EDTA (0.05%) was purchased from Gibco-BRL (Grand Island, NY). Anti-HMGCS2 and anti-TBP antibodies were purchased from Abcam (Cambridge, UK). Anti-NF-kB was purchased from Cell Signaling Technology (Danvers, MS). Anti-ANGPTL3 antibody and secondary antibodies (goat anti-mouse and rabbit IgG-horseradish peroxidase) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The WesternBright ECL HRP substrate detection solution was purchased from Advansta Inc. (Menlo Park, CA). The NE-PER kit and Restore™ Western Blot Stripping Buffer were purchased from Pierce (Rockford, IL). QIAprep Spin Miniprep Kit and RNeasy Mini Kit were purchased from Qiagen (Hilden, Germany). RT-PCR Premix kit was obtained from Bioneer (Daejon, Korea). Primers specific for amplifying HMGCS2, NF-kB, and 18s RNA transcripts were purchased from Integrated DNA Technologies (Coralville, IA). Oligonucleotide probes (NF-kB) and the electrophoretic mobility shift assay (EMSA) kit were obtained from Promega Corp (Madison, WI). NF-kB inhibitor, sodium 4-aminosalicylate (4-ASA), was purchased from Selleckchem (Houston, TX).
Cell culture and treatment
Mouse normal hepatocyte cell line (FL83B) were purchased from ATCC and maintained in F-12K medium containing 10% FBS and 100 U/ml penicillin and streptomycin at 37°C in 5% CO2. The cells were placed in airtight chambers (NuAire, Plymouth, MN). At the beginning of each experiment, the cells were re-suspended in F-12K medium at a density of 2.5 × 105 cells/ml. Cells were treated with D-glucose and silibinin (40 μM) for 24 h.
Cell proliferation inhibition
Cell viability was assayed by adding 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) and measuring the blue formazan produced. MTT is metabolized by mitochondrial dehydrogenase, which is active only in live cells. The cells were re-suspended in medium one day before drug treatment, at a density of 3 × 103 cells per well in 96-well culture plates. To determine growth inhibition, cells were incubated with various concentrations of silibinin. For control cells, the medium was replaced with media with DMSO as vehicle control. To measure cell viability, MTT (5 mg/ml) was added to each well and incubated for 4 h at 37°C. The formazan product formed was dissolved by adding 200 μl dimethyl sulfoxide (DMSO) to each well, and the absorbance was measured at 550 nm on an Ultra Multifunctional Microplate Reader (TECAN, Durham, NC). All measurements were performed in triplicate, and experiments were repeated at least three times.
β-Hydroxybutyrate colorimetric assay
Measurements of BHB were performed with a colorimetric assay kit, according to the manufacture’s protocol. Briefly, FL83B cells (~ 18 × 106) were collected by centrifugation and sonicated in cold diluted assay buffer. The supernatant was removed, and cells were re-suspended in cold diluted assay buffer. BHB standard wells and sample wells were prepared as per the protocol. The reaction was initiated by adding developer solution. Cells were incubated at 25°C in the dark for 30 min; then, absorbance was measured at 450 nm.
Western blotting
FL83B cell lines were treated with high glucose (3.78 g/l; 0.37%) and silibinin (40 μM) for pre-determined times. Whole cell cultures were lysed on ice in radio immune precipitation (RIPA) lysis buffer, which contained phosphatase and protease inhibitors. Cells were centrifuged at 15,000 rpm for 10 min at 4°C to remove cellular debris. Protein concentrations were measured with the Bradford method. Western blotting analysis was performed to detect HMGCS2, NF-kB, ANGPTL3, TBP, and β-actin expression. Equal amounts of proteins were resolved on SDS-PAGE and transferred onto nitrocellulose membranes. The blots were blocked for 1 h with 5% skim milk. The membrane was probed overnight at 4°C with primary antibodies. Detection with HRP-conjugated secondary antibodies was performed with the WesternBright ECL HRP substrate detection kit and a LAS-4000 imaging device (Fujifilm, Japan).
Reverse transcription polymerase chain reaction
FL83B cell lines were treated with high glucose (3.78 g/l; 0.37%) and silibinin (40 μM) for 24 h. Total RNA was extracted with the RNeasy Mini Kit (Qiagen) and quantified spectrophotometrically at 260 nm. We performed reverse-transcription and PCR (RT-PCR) analyses to detect expression of HMGCS2, NF-kB, and 18s RNA. First, cDNA was synthesized from total RNA with a first-strand cDNA synthesis kit (Bioneer, Korea). Reverse transcription was performed at 42°C for 1 h and 80°C for 15 min. PCR was conducted with the cDNA. PCR products were analyzed on a 1% agarose gel stained with ethidium bromide.
Electrophoretic mobility shift assay
The DNA-binding activity of NF-kB was detected with an electrophoretic mobility shift assay (EMSA). Briefly, nuclear protein extracts were prepared from treated and untreated cells with the Nuclear Extract Kit (Panomics, Fremont, CA; AY2002). The EMSA experiment was performed by incubating a biotin-labeled transcription factor (TF-NF-kB probe) with the nuclear extracts. Proteins were resolved on a non-denaturing 6% PAGE gel (Bio-Rad, Korea). The proteins in the gel were transferred to a nylon membrane, and the TF-NF-kB probe was detected with streptavidin-HRP and a chemiluminescent substrate.
NF-kB over-expression analysis
FL83B cells (1 × 105) were cultured in 6-well plates and grown to 60% confluence. The cells were then transfected with wild-NF-kB vector (cFlag pcDNA3) with the DharmaFECT transfection reagent (Dharmacon, USA) for 24 h. Transfected cells were then treated for an additional 24 h with low glucose (1.26 g/l; 0.12%) media that contained 40 μM silibinin. Proteins were isolated and analyzed with Western blotting to determine NF-kB and HMGCS2 expression levels.
Statistical analysis
All the experiments were repeated three times, and the results were expressed as mean ± SEM. Groups were compared with the Student’s t test. Statistical analyses were performed with the SAS program.
Results
Silibinin inhibits proliferation of FL83B mouse hepatocytes
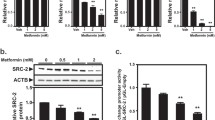
We used an MTT assay to analyze the inhibitory effects of silibinin on FL83B mouse hepatocyte proliferation (Fig. 1A). We aimed to determine the concentration of silibinin that would produce anti-ketotic activity with little or no effect on cell growth. We analyzed FL83B cell proliferation at increasing concentrations of silibinin. The results showed high percentages of live cells at low concentrations of silibinin, and 40 μM silibinin caused little cell death (18%). Therefore, we used 40 μM for further experiments.
Silibinin inhibited FL83B mouse hepatocyte proliferation and ketosis in vitro. (A) MTT results show that silibinin inhibits FL83B cell proliferation in a dose-dependent manner. The minor effect of 40 μM silibinin indicated that it could be used in further analysis. (B) Colorimetric quantification of β-hydroxybutyrate (BHB) shows the in vitro ketosis in high glucose (3.78 g/l; 0.37%) and in low glucose (1.26 g/l; 0.12%) conditions, with or without 40 μM silibinin. (C) Graphical representation of relative BHB levels in FL83B cells treated in media with high glucose and in low glucose, with or without 40 μM silibinin. ***P < 0.001, based on the Student t test.
Silibinin inhibited in vitro ketosis in FL83B cells
We established in vitro ketosis conditions by applying glucose starvation to FL83B cells. We checked BHB expression levels to determine when ketosis was induced. A comparison of BHB expression levels between cells treated with low glucose (1.26 g/l; 0.12%) and high glucose (3.78 g/l; 0.37%) showed that glucose starvation successfully induced ketosis (Fig. 1B, C). We checked the anti-ketotic effects of silibinin-treating ketotic FL83 cells with 40 μM silibinin. The results showed that silibinin reduced BHB levels compared to the levels observed in ketosis. The inhibition of ketone body production suggested that our candidate drug had anti-ketotic properties.
Silibinin inhibited ketosis by regulating HMGCS2 through NF-kB signaling
HMGCS2 contributes to beta oxidation, which leads to ketone body formation. NF-kB is a transcription factor for HMGCS2. Therefore, an effective anti-ketosis drug should aim to down-regulate the expression levels of both HMGCS2 and NF-kB. In this study, we tested whether 40 μM silibinin (the candidate drug) could down-regulate the expression levels of HMGCS2 and NF-kB. We divided cells into three treatment groups: the low glucose (1.26 g/l; 0.12%) group (ketosis group), the high glucose (3.78 g/l; 0.37%) group (positive control), and the low glucose group treated with 40 μM silibinin. We isolated total proteins and total RNA for analysis. RTPCR analyses of total RNA showed that 40 μM silibinin down-regulated both HMGCS2 and NF-kB expression (Fig. 2A). Silibinin significantly inhibited HMGCS2 expression (Fig. 2B). Total proteins analyzed on western blots (Fig. 2C, D) showed the same results; thus, silibinin had anti-ketotic effects at both the RNA and protein levels. Because angiopoietin-like 3 (ANGPTL3) has been considered a molecular target for ketosis treatment, we also tested whether silibinin affected angiopoietin-like 3 (ANGPTL3) expression. We found that silibinin inhibited ANGPTL3 expression, which provided strong support for silibinin as an anti-ketosis agent (Fig. 2C).
Silibinin inhibited ketosis by regulating HMGCS2 and NF-kB signaling molecules. (A) RT-PCR results show HMGCS2 and NF-kB RNA expression after cells were treated with 40 μM silibinin for 24 h. (B) Graphical representation of HMGCS2 and NF-kB inhibition by silibinin at the transcriptional level. (C) Western blot results shows the expression patterns of ANGPTL3, HMGCS2, and NF-kB proteins, after cells were treated with 40 μM silibinin for 24 h. (D) Relative expression of ANGPTL3, HMGCS2, and NF-kB proteins. *P < 0.05, **P < 0.01, and ***P < 0.001, based on the Student t test.
Silibinin inhibited the nuclear translocation and DNA binding activity of NF-kB in FL83B mouse hepatocytes
Based on our finding that silibinin inhibited NF-kB expression at the transcriptional and translational levels, we reasoned that silibinin might influence NF-kB nuclear translocation, which is required for the transcriptional role of NF-kB in the nucleus. Therefore, we analyzed nuclear extracts isolated from cells grown in low glucose (1.26 g/l; 0.12%), high glucose (3.78 g/l; 0.37%), and low glucose with silibinin (Fig. 3A). We found that silibinin significantly inhibited the expression of HMGCS2 and NF-kB in nuclear extracts (Fig. 3B). These results suggested that silibinin inhibited the nuclear translocation of both NF-kB and HMGCS2. Next, we analyzed the DNA binding activity of NF-kB with an electrophoretic mobility shift assay (EMSA). We used a labeled DNA oligonucleotide (gamma interferon activation site), which was known to bind specifically to NF-kB, to examine the DNA binding activity. We found that silibinin down-regulated the formation of the NF-kB/DNA complex (Fig. 3C). These results suggested that NF-kB might be involved in the anti-ketosis effects of silibinin.
Silibinin enhanced the nuclear translocation and DNA-binding activity of NF-kB in FL83B cells. (A) Western blot analysis of nuclear extract proteins from cells exposed to low glucose (1.26 g/l; 0.12%), high glucose (3.78 g/l; 0.37%), or low glucose plus 40 μM silibinin for 24 h. (B) Relative expression patterns of HMGCS2 and NF-kB nuclear proteins. (C) Gel shift analysis of nuclear extracts shows NF-kB binding to the gamma interferon activation site element in DNA from cells treated with low glucose, high glucose, or low glucose plus 40 μM silibinin. Images are representative of three independent experiments. **P < 0.01 and ***P < 0.001, based on the Student t test.
NF-kB positively regulated the expression of HMGCS2 in ketosis treatment
Silibinin inhibited NF-kB expression and its DNA binding activity. Silibinin also inhibited the expression of HMGCS2 at the RNA and protein levels. These results indicated that HMGCS2 and NF-kB were inhibited upon silibinin treatment. To determine the exact relationship between NF-kB and HMGCS2, we inhibited NF-kB activity with a specific inhibitor, sodium 4-aminosalicylate (4-ASA). First, we examined the down-regulation of NF-kB expression at different concentrations of 4-ASA with western blotting. We found that 0.5 mM 4-ASA was appropriate for inhibiting NF-kB expression (Fig. 4A). We also found that this NF-kB inhibitor also inhibited the expression of HMGCS2 in whole cell lysates. Next, we performed western blotting to analyze total protein expression with or without 4-ASA (Fig. 4B). In 4-ASA-treated cells, silibinin significantly down-regulated the expression of HMGCS2, compared to cells that were not exposed to silibinin (Fig. 4C). Nevertheless, down-regulation of HMGCS2 expression by the NF-kB inhibitor suggested that NF-kB acted as a positive regulator for HMGCS2. This relationship was confirmed when we over-expressed NF-kB in FL83B cells by transfecting with a vector that carried the wild-type NF-kB DNA sequence. A western blot analysis of HMGCS2 expression (Fig. 5A) showed an increase in the expression of HMGCS2, in both silibinin-treated and silibinin-treated cells (Fig. 5B). These results showed that NF-kB played a vital role in the effect of silibinin on ketosis by acting as a positive regulator of HMGCS2.
Role of NF-kB in the expression of HMGCS2 under ketotic conditions. (A) Western blot analysis of HMGCS2 and NF-kB in cells treated with different concentrations of the NF-kB inhibitor, 4-ASA. (B) Western blot analysis shows the results of NF-kB inhibition with 4-ASA, and the levels of HMGCS2 and NF-kB in cells treated with silibinin for 24 h. (C) Graphical representation of the relative expression of NF-kB and HMGCS2 with respect to β-actin. *P < 0.05, based on the Student t test.
NF-kB effects on the expression of HMGCS2 during anti-ketosis treatment. (A) Western blot analysis shows the expression of HMGCS2 and NF-kB after cells that over-expressed NF-kB were treated with silibinin for 24 h. (B) Relative expression levels of NF-kB and HMGCS2 compared to β-actin. **P < 0.01 and ***P < 0.001, based on the Student t test.
Discussion
The induction of in vitro ketosis with the glucose starvation method can be used to identify anti-ketosis effects of a candidate drug (Sreekantan Preetha et al. 2015). Here, we used the low glucose concentration (0.12%) media (F-12K) to induce ketosis, and we increased the glucose concentration thrice for control conditions (high-glucose condition; 3.78 g/l; 0.37% glucose). We showed that the BHB level was significantly reduced in the high glucose group compared to the low glucose (1.26 g/l; 0.12%) group (Fig. 2B). Silibinin is a flavonoid that has been used as a hepatoprotector for several years (Hogan et al. 2007). It is typically given orally to treat hepatic diseases (Cheung et al. 2010). Silibinin is very active in liver, and ketosis is produced in the liver; therefore, we tested silibinin activity against ketosis in FL83B mouse hepatocytes. Our results showed that silibinin reduced the BHB levels in the group of cells treated with low glucose. At the same time, the concentration of silibinin is considered an important factor in anti-ketosis treatments. An anti-ketosis drug should be designed to cause little or no effect on cell growth. We found that a concentration of 40 μM silibinin inhibited 18% of cell proliferation, after a treatment period of 24 h. Our BHB analysis showed that 40 μM silibinin could inhibit ketosis in vitro.
HMGCS2 and ANGPTL3 promote ketosis through different pathways. During beta oxidation, HMGCS2 plays an important role in the conversion of acetyl-CoA and acetoacetyl-CoA to HMG-CoA, which is then converted to acetoacetic acid and BHB (Vilà-Brau et al. 2011). In contrast, ANGPTL3 is involved in carbohydrate and fat metabolism disorders; thus, it is considered a molecular target for ketosis treatments (Xiao et al. 2012). Inhibition of these molecules leads to the inhibition of ketosis. Silibinin inhibited both these target molecules at the transcriptional level, and it also inhibited HMGCS2 at the translational level, in FL83B cells. These results indicated that silibinin had anti-ketotic properties. In addition, silibinin blocked the nuclear translocation of HMGCS2. Taken together, these findings showed that silibinin inhibited ketosis by blocking the expression and activity of HMGCS2 during beta oxidation.
NF-kB is a transcription factor that regulates the HMGCS2 gene. The NF-kB signaling pathway participates in the production of CCL and CXCL (Nookala et al. 2013), and CXCL chemokines are inhibited by PPARα activators (Antonelli et al. 2010). HMGCS2 interacts with PPARα to co-activate PPRE sites in DNA to increase gene transcription (Kostiuk et al. 2010). Thus, NF-kB plays a role in the activity of HMGCS2 through various signaling molecules. However, it was unknown whether NF-kB played role in the expression of HMGCS2 during ketosis treatment. Recent studies showed that NF-kB played a vital role in osteoclastogenesis in bone marrow–derived macrophages (Joung et al. 2016). A previous study showed that elevated BHB concentrations activated the NF-kB signaling pathway, and thereby induced hepatocyte inflammatory disorder (Shi et al. 2014), which suggested that NF-kB might play a role in ketosis. We investigated the role of NF-kB in ketotic FL83B cells. Our candidate drug, silibinin, reduced NF-kB expression at the transcriptional level and the translational level. Additionally, 40 μM silibinin suppressed the nuclear translocation and the DNA binding activity of NF-kB. These results showed that ketosis treatment down-regulated NF-kB expression, which suggested that NF-kB played a role in ketosis.
We found that HMGCS2 and NF-kB played vital roles in ketosis. Thus, these molecules might be considered molecular targets for treating ketosis. Although NF-kB is a transcription factor for HMGCS2, it remained unknown whether this regulation was positive or negative. Our results showed that silibinin treatment down-regulated the expression of both molecules. Therefore, we hypothesized that NF-kB acted as a positive regulator of HMGCS2. To confirm this, we inhibited NF-kB with its specific inhibitor and then checked the expression of HMGCS2. The result showed that HMGCS2 expression was also down-regulated in NF-kB inhibitor–treated cells. Conversely, NF-kB over-expression up-regulated HMGCS2 expression. These results provided solid evidence that NF-kB acted as a positive regulator of HMGCS2.
Conclusion
In conclusion, this study showed that silibinin inhibited in vitro ketosis by inhibiting an HMGCS2-targeted NF-kB signaling pathway and by inhibiting the DNA binding activity of NF-kB. Our study also showed that NF-kB acted as a positive regulator of HMGCS2 during ketotic conditions. Altogether, silibinin can be considered as a candidate drug for anti-ketosis research and the added advantage of silibinin as a drug is it has less side effects and more efficacy.
References
Abrahan C, Ash JD (2016) The potential use of PGC-1α and PGC-1β to protect the retina by stimulating mitochondrial repair. Adv Exp Med Biol 854:403–409
Antonelli A, Ferrari SM, Frascerra S, Pupilli C, Mancusi C, Metelli MR, Orlando C, Ferrannini E, Fallahi P (2010) CXCL9 and CXCL11 chemokines modulation by peroxisome proliferator-activated receptor-alpha agonists secretion in Graves’ and normal thyrocytes. J Clin Endocrinol Metab 95:E413–E420
Cheung CW, Gibbons N, Johnson DW, Nicol DL (2010) Silibinin—a promising new treatment for cancer. Anti Cancer Agents Med Chem 10:186–195
Cotter DG, Ercal B, Huang X, Leid JM, d'Avignon DA, Graham MJ, Dietzen DJ, Brunt EM, Patti GJ, Crawford PA (2014) Ketogenesis prevents diet-induced fatty liver injury and hyperglycemia. J Clin Invest 124:5175–5190
Davis-Searles PR, Nakanishi Y, Kim NC, Graf TN, Oberlies NH, Wani MC, Wall ME, Agarwal R, Kroll DJ (2005) Milk thistle and prostate cancer: differential effects of pure flavonolignans from Silybum marianum on antiproliferative end points in human prostate carcinoma cells. Cancer Res 65:4448–4457
Duyao MP, Buckler AJ, Sonenshein GE (1990) Interaction of an NF-kappa B-like factor with a site upstream of the c-myc promoter. Proc Natl Acad Sci U S A 87:4727–4731
Garber AJ, Menzel PH, Boden G, Owen OE (1974) Hepatic ketogenesis and gluconeogenesis in humans. J Clin Invest 54:981–989
Hegardt FG (1999) Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem J 338:569–582
Hogan FS, Krishnegowda NK, Mikhailova M, Kahlenberg MS (2007) Flavonoid, silibinin, inhibits proliferation and promotes cell-cycle arrest of human colon cancer. J Surg Res 143:58–65
Iademarco MF, McQuillan JJ, Rosen GD, Dean DC (1992) Characterization of the promoter for vascular cell adhesion molecule-1 (VCAM-1). J Biol Chem 267:16323–16329
Joung YH, Darvin P, Kang DY, Sp N, Byun HJ, Lee CH, Lee HK, Yang YM (2016) Methylsulfonylmethane inhibits RANKL-induced osteoclastogenesis in BMMs by suppressing NF-κB and STAT3 activities. PLoS One 11:e0159891
Kostiuk MA, Keller BO, Berthiaume LG (2010) Palmitoylation of ketogenic enzyme HMGCS2 enhances its interaction with PPARalpha and transcription at the Hmgcs2 PPRE. FASEB J 24:1914–1924
Kumar S, Raina K, Agarwal C, Agarwal R (2014) Silibinin strongly inhibits the growth kinetics of colon cancer stem cell-enriched spheroids by modulating interleukin 4/6-mediated survival signals. Oncotarget 5:4972–4989
Lawrence T (2009) The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol 1:a001651
Mateen S, Tyagi A, Agarwal C, Singh RP, Agarwal R (2010) Silibinin inhibits human nonsmall cell lung cancer cell growth through cell-cycle arrest by modulating expression and function of key cell-cycle regulators. Mol Carcinog 49:247–258
Nookala AR, Shah A, Noel RJ, Kumar A (2013) HIV-1 tat-mediated induction of CCL5 in astrocytes involves NF-κB, AP-1, C/EBPα and C/EBPγ transcription factors and JAK, PI3K/Akt and p38 MAPK signaling pathways. PLoS One 8:e78855
Saller R, Brignoli R, Melzer J, Meier R (2008) An updated systematic review with meta-analysis for the clinical evidence of silymarin. Forsch Komplementmed 15:9–20
Shi X, Li X, Li D, Li Y, Song Y, Deng Q, Wang J, Zhang Y, Ding H, Yin L, Zhang Y, Wang Z, Li X, Liu G (2014) β-Hydroxybutyrate activates the NF-κB signaling pathway to promote the expression of pro-inflammatory factors in calf hepatocytes. Cell Physiol Biochem 33:920–932
Singh RP, Agarwal R (2005) Mechanisms and preclinical efficacy of silibinin in preventing skin cancer. Eur J Cancer 41:1969–1979
Sreekantan Preetha N, Kang DY, Darvin P, Kim DN, Joung YH, Kim SY, Cho KH, Do CH, Park KD, Lee JH, Lee HY, Park JH, Yang YM (2015) Induction of in vitro ketosis condition and suppression using methylsulfonylmethane by altering ANGPTL3 expression through STAT5b signaling mechanism. Anim Cells Syst 19:30–38
Tensen CP, Flier J, Rampersad SS, Sampat-Sardjoepersad S, Scheper RJ, Boorsma DM, Willemze R (1999) Genomic organization, sequence and transcriptional regulation of the human CXCL 11(1) gene. BiochimBiophysActa 1446:167–172
Ueda A, Okuda K, Ohno S, Shirai A, Igarashi T, Matsunaga K, Fukushima J, Kawamoto S, Ishigatsubo Y, Okubo T (1994) NF-kappa B and Sp1 regulate transcription of the human monocyte chemoattractant protein-1 gene. J Immunol 153:2052–2063
Vilà-Brau A, De Sousa-Coelho AL, Mayordomo C, Haro D, Marrero PF (2011) Human HMGCS2 regulates mitochondrial fatty acid oxidation and FGF21 expression in HepG2 cell line. J Biol Chem 286:20423–20430
Wang HJ, Wei XF, Jiang YY, Huang H, Yang Y, Fan SM, Zang LH, Tashiro S, Onodera S, Ikejima T (2010) Silibinin induces protective superoxide generation in human breast cancer MCF-7 cells. Free Radic Res 44:90–100
Xiao HB, Sun ZL, Zhou N (2012) 1,3,5,8-Tetrahydroxyxanthone regulates ANGPTL3-LPL pathway to lessen the ketosis in mice. Eur J Pharm Sci 46:26–31
Zeng J, Sun Y, Wu K, Li L, Zhang G, Yang Z, Wang Z, Zhang D, Xue Y, Chen Y, Zhu G, Wang X, He D (2011) Chemopreventive and chemotherapeutic effects of intravesicalsilibinin against bladder cancer by acting on mitochondria. Mol Cancer Ther 10:104–116
Funding
This work was supported by a grant from the Next Generation BioGreen 21 Program (No. PJ01110901, PJ01315101), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Editor: Tetsuji Okamoto
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kang, D.Y., Sp, N., Do Park, K. et al. Silibinin inhibits in vitro ketosis by regulating HMGCS2 and NF-kB: elucidation of signaling molecule relationship under ketotic conditions. In Vitro Cell.Dev.Biol.-Animal 55, 368–375 (2019). https://doi.org/10.1007/s11626-019-00351-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-019-00351-6