Abstract
Prostate stromal cells may play binary roles in the process of prostate cancer development. As the first to be encountered by infiltrating prostate cancer cells, prostate stromal cells form the first defense line against prostate cancer progression and metastasis. However, interaction between prostate cancer and stromal cells may facilitate the formation of a tumor microenvironment favoring cancer cell growth and survival. To establish an experimental system for studying the interaction between cancer and stromal cells, we isolated three matched pairs of normal and cancer-associated human prostate stromal clones. In this report, we describe the morphologic and behavioral characteristics of these cells and their effect on LNCaP prostate cancer cells in co-culture. Unlike LNCaP prostate cancer cells, the isolated prostate stromal clones are large fibroblast-like cells with a slow proliferation rate. Growth and survival of these clones are not affected by androgens. The stromal cells display high resistance to serum starvation, while cancer-associated stromal clones have differentiated survival ability. In co-culture experiments, the stromal cells protected some LNCaP prostate cancer cells from death by serum starvation, and cancer-associated stromal clones showed more protection. This work thus established a panel of valuable human prostate stromal cell lines, which could be used in co-culture to study the interaction between prostate cancer and prostate stromal cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The prostate stroma is involved in prostate oncogenesis and prostate cancer progression (Chung 1995; Harding and Theodorescu 2000; Sung and Chung 2002; Condon 2005; Loberg et al. 2005). Completely embedding the epithelial glandular branches, the prostate stroma is predictably the first zone encountered by cancer cells breaking away from the epithelial layer. Whether the stromal compartment inhibits or promotes prostate cancer progression would have a deep impact on the entire process of the disease.
The stromal compartment is an integral part of the functional prostate. During embryonic development, the stromal cells direct budding and ductal branching morphogenesis (Chung and Cunha 1983; Cunha 1994; Cunha et al. 1995, 2004). In tissue reconstitution studies, embryonic stromal cells determine the fate of the epithelial differentiation (Chung and Cunha 1983; Cunha 1994; Chung 1995; Cunha et al. 1995, 2002, 2004). Mechanistically, it is suggested that prostate stromal cells respond to sex hormones by producing andromedins, soluble factors mediating growth, and differentiation of the epithelial compartment (Yan et al. 1992; Fasciana et al. 1996; Lu et al. 1999; Planz et al. 1999). In prostate cancer, the functional characteristics of the prostate stroma could be hijacked by cancer cells to form a favorable tumor microenvironment. We have demonstrated that stromal cells in the tumor microenvironment play an important role in determining the fate of cancer cells. Through chimeric tumor formation with MS osteosarcoma cells, for instance, androgen-dependent and non-tumorigenic LNCaP prostate cancer cells can be converted to the androgen-independent and tumorigenic C4-2 subline (Gleave et al. 1991; Wu et al. 1994). Similar changes were observed in LNCaP cells after being co-cultured with MG63 osteosarcoma cells (Rhee et al. 2001). In addition, by xenograft tumor formation and metastasis, the epithelial cell-like ARCaPE prostate cancer cells could be changed into mesenchymal stroma-like cells displaying increased tumorigenicity and metastatic potency (Xu et al. 2006; He et al. 2009). These studies established that interaction with stromal cells is a critical step for cancer cells to acquire additional malignant potential during prostate cancer progression and metastasis.
It should be pointed out that many studies on cancer–stromal interaction take osteosarcoma or immortalized stromal cells as surrogates for mesenchymal stromal cells. Xenograft tumor formation in athymic mice may not be easily adapted to mechanistic studies of cancer–stromal interaction. Although prostate stromal cells are suspected as a culprit in prostate oncogenesis, there is little direct evidence pinpointing a causal role for these cells, and the pathologic relevance of stromal cells to prostate cancer remains to be confirmed. A major hindrance to the study of prostate stromal cells is the lack of an experimental system in which well-characterized stromal cells can be studied in a reproducible fashion, and their role examined by simulating the interaction between cancer and stromal cells.
We have established three matched pairs of normal and cancer-associated human prostate stromal cell lines to facilitate the study of cancer–stromal interaction. In this report, we outline the isolation and initial characterization of these cell lines. Their role in promoting prostate cancer progression is exemplified by a study of cancer–stromal interaction in a co-culture of prostate cancer and stromal cells.
Materials and Methods
Human prostate cancer cell lines.
Unless specified, all the cells used in this study were maintained in T-medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS, Atlanta Biologicals, Lawrenceville, GA), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C, in humidified atmospheric air supplemented with 5% CO2. The human prostate cancer cell line LNCaP used was a kind gift of Dr. Gary Miller (University of Colorado, Denver, CO) (Gleave et al. 1991). For this study, cells between original passages 29 and 34 were used.
Cloning prostate stromal cells from clinical specimens.
Three matched pairs of normal and cancer-associated prostate specimens were obtained under an IRB protocol. The specimens were from three prostate cancer patients who underwent radical prostatectomy in the Department of Urology, Emory University School of Medicine. No identifier or disease history was provided, but the tumor in these prostates was confirmed by histopathologic diagnosis. To obtain a normal prostate specimen, a cube (<150 mm3) of prostate tissue was dissected by a pathologist from a histologically normal region distal to the tumor. Another cube was dissected from a cancer-affected zone from the same prostate to provide a matched cancer specimen.
Upon arrival at the laboratory, the specimens were diced with a sterile razor blade. The sample was incubated at 37°C for 1 h in phosphate-buffered saline (PBS) containing 2.4 units of Dispase II (Roche Diagnostics, Indianapolis, IN). After washing in PBS, live cells were plated in low density (1 × 105) onto a 15-cm culture dish for 2 wk. Epithelial cells with low attachment to plastic surface were collected in the medium for further characterization. The culture dish was rinsed three times before picking stromal colonies, which were tightly attached to the dish. For each specimen, 12 colonies were picked with cloning disk and amplified. A representative clone was used in this study. For comparison, PrSC, a primary normal human prostate stromal cell line, was purchased from Lonza Walkersville, Inc. (Walkersville, MD). PrSC cells between passages 6 and 11 cultured in T medium were used.
Serum starvation and androgen deprivation.
Cells (2 × 105/well) were plated onto six-well plates. After 24 h for attachment, the culture was rinsed with PBS to remove serum. Serum starvation was carried out by culturing cells with serum-starvation medium, which was phenol red-free RPMI 1640 medium (Invitrogen) containing no FBS. To perform androgen deprivation, cells were cultured first in serum-starvation medium for 48 h to exhaust androgens and then in androgen-deprivation medium, which was phenol red-free RPMI 1640 medium containing 1% charcoal/dextran stripped FBS (Hyclone, Logan, UT).
Androgen stimulation and measurement of PSA.
Cells on six-well plates at 75% confluence were first subjected to 48-h serum starvation and then treated with 5 nM of synthetic androgen methyltrienolone (R1881, Perkin Elmer, Waltham, MA) in the androgen-deprivation medium. Culture medium was collected for measuring prostate-specific antigen (PSA) concentration with commercial PSA ELISA kits (United Biotech Inc., Mountain View, CA). Triplicate assays were performed on each sample.
Transfection.
The protocol for transfecting prostate cancer cells with linearized pAsRed2 eukaryotic expression vector was previously reported (He et al. 2009). In this study, LNCaP cells were used in transfection. G418 (300 μg/ml) was used to select for stably transfected cells, which were cloned subsequently by the method of limited dilution. RL-1, a selected LNCaP clone stably expressing the AsRed2 red fluorescence protein, was used in this study.
Cancer–stromal co-culture.
Prostate stromal cells were grown in six-well plates with 2 ml culture medium in each well. The cells were allowed to grow to full confluence. After removing culture medium from the stromal cells, prostate cancer LNCaP cells in low density (5 × 104/ml) in 4 ml fresh medium were overlaid onto the stromal monolayer. The volume of the medium in co-culture was always double the volume used when each cell type was cultured alone.
Cell proliferation assay.
Crystal violet staining was used to determine the growth rate using our published protocol (Xu et al. 2006). In this study, cells from each stromal cell line were plated in low density (1 × 104/well) onto 24-well plates in triplicate wells. The culture was terminated 4 d later, before any of the cell lines reached confluence. Following crystal violet staining, relative growth rate was determined by spectrophotometric reading at OD595.
Colony formation assay.
Cells in six-well plates were subjected to serum starvation in triplicate for 60 d. The cells in each well were then collected as single-cell suspension and plated onto a 15-cm dish. After cultured in T medium containing 10% FBS for 2 wk, colonies in each dish were counted. To assay colony formation of co-cultured cancer and stromal cells, red fluorescence microscopy was used to distinguish colonies of the red fluorescent prostate cancer cells from colonies of stromal cells.
Microphotography.
The equipment and software used in phase contrast and fluorescence microphotography was reported previously (Xu et al. 2006; He et al. 2009). Cell size was determined with the Image-Pro Plus software, which was calibrated with a micrometer.
Statistical analysis.
Student’s t test was used. Level of statistical significance between control and treated groups was set at P < 0.05.
Results
Establishment of matched pairs of human prostate stromal cell lines.
We obtained three matched pairs of normal and cancer-associated human prostate specimens. After the specimens were disintegrated into single-cell preparations, cells in each preparation were plated in low density and cultured for colony formation. From each ex vivo culture, 12 stromal cell-like colonies tightly attached to the culture dish were picked and amplified as individual clones. By inspection during the first three passages, we found that all clones from each specimen had a similar cellular morphology and growth rate. A representative clone from each specimen was used for further investigation.
Using this protocol, we established three matched pairs of human prostate stromal cell clones. These clones were named as human prostate stromal (HPS) cells, with a numerical affix reflecting the chronological order of the isolation. These paired clones are HPS-10 (normal) and HPS-11 (cancer-associated), HPS-12 (normal) and HPS-13 (cancer-associated), and HPS-14 (normal) and HPS-15 (cancer-associated).
Characteristics of the prostate stromal cell clones.
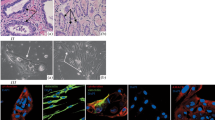
On microscopic inspection, the most prominent feature of the prostate stromal clones is their expanded cell size. Whereas the PrSC normal prostate stromal fibroblast cells are large, the HPS clones all showed even larger cell sizes (Fig. 1). At sub-confluence, an average HPS cell measured 300 × 50 μm at the largest dimensions. On the other hand, the HPS cells were flatter than cancer cells, yielding less refraction during phase contrast microscopy.
The normal and cancer-associated stromal counterparts shared cellular morphology and size, but each matched pair could be different from the other pairs (Fig. 1). This is especially obvious for the pair of HPS-12 and HPS-13. Both the normal HPS-12 and the cancer-associated HPS-13 stromal cells shared an expanded and flat shape, almost indistinguishable from each other. On the other hand, this pair of cells could be easily distinguished from other matched pairs.
We tested whether cells of the HPS clones could endure prolonged culture without entering senescence. Using a quarter of the first 1 × 106 cells as passage 1, we cultured the matched pairs for 60 passages, with a 1:3 subculture ratio. Whereas changes in growth rate were minimal during the first 30 passages, proliferation of the HPS clones declined along with the passage, and growth was halted in many clones beyond 45 passages. Eventually, only one matched pair (HPS-14 and HPS-15) and another cancer-associated stromal clone (HPS-11) reached passage 60. It appeared that these established clones represented stable primary prostate stromal cell lines, which could be continuously cultured for experimental use for less than 30 passages.
The HPS clones all showed low proliferation activity. When examined for growth rate in sub-confluence, all the clones showed even slower growth than the original LNCaP prostate cancer cell line (Fig. 2 A), which is known to have a slow growth rate (Singh et al. 1999). Interestingly, we did not observe a differential proliferation in cancer-associated stromal cells, since none of the cancer-associated stromal clones grew faster than its normal counterpart.
Androgen-independent growth of the HPS cell clones. A HPS cells were treated with androgen-deprivation for 96 h and assayed for cell proliferation by crystal violet staining. B HPS cells were treated with synthetic androgen R1881 for 96 h and assayed for cell proliferation. In these experiments, growth of the androgen-dependent human LNCaP prostate cancer cells was used as a positive control. Data represent the mean of a triplicate assay, and the results are representative of two separate experiments. For each cell line, an asterisk indicates statistical significance (p < 0.05) compared to control group of the cell line.
Prostate stromal cells are insensitive to androgen deprivation.
Prostate stromal cells may have differentiated sensitivity to androgens compared to prostate epithelial cells. As androgen deprivation in vivo effectively causes programmed death of the epithelial cells, it is less effective on the stromal compartment (English et al. 1985; Sagalowsky 1985; Lippert and Keefer 1987; McConnell 1990; Sensibar et al. 1990). To assess whether the established HPS clones inherited the same property, we cultured the cells in androgen deprivation medium for 4 d and determined the growth rate. Compared to the LNCaP cells whose growth was inhibited under these conditions, the HPS clones grew well. No significant changes in growth rate were detected in the treated cells (Fig. 2 A). In addition, we determined that growth of the established stromal cells was not stimulated by the synthetic androgen R1881 (Fig. 2 B). It seemed that proliferation of the established prostate stromal clones proceeded in an androgen-independent fashion.
To determine the survival potential of the stromal cells, we compared these cells with LNCaP prostate cancer cells for their survival after serum starvation. Under these culture conditions, stromal cells maintained the same morphology for more than 4 wk. Subsequently, the cells became narrower while some detached and were removed by change of medium. The majority of the cells survived and the stromal monolayer remained intact (Fig. 3 A). In two experiments, to maintain cells for 60 d under androgen-free and serum-free conditions, all the stromal clones survived the experiments. At the end of the 60-d treatment, the vitality of the cells was confirmed by colony formation assays, in which recovered cells formed substantial numbers of colonies (Fig. 3 B). In parallel experiments, the androgen-dependent LNCaP prostate cancer cells failed to survive serum starvation. Most died around 5 wk, and by 60 d, no LNCaP colonies were formed in two colony formation experiments. Relative to LNCaP prostate cancer cells, the established prostate stromal clones were highly resistant to androgen deprivation and serum starvation.
Prostate stromal cells are resistant to serum starvation. A Morphologies of a monolayer of HPS-15 cells at the beginning (Day 1) and end (Day 60) of serum starvation. The starved cells became narrower, attached by abundant extracellular matrix materials. B after 60 d of serum starvation, surviving stromal cells were subjected to colony formation for 2 wk. Two representative colonies are shown from the surviving HPS-15 cells. C Results of the colony formation by surviving stromal cells after 60 d of serum starvation. Data represent the mean of triplicate assays. For each matched pair, an asterisk indicates statistical significance (p < 0.05) compared to the counterpart.
In these studies, when matched normal and cancer-associated stromal cells were compared for their survival capability, two cancer-associated stromal clones (HPS-11 and HPS-15) formed significantly more colonies than their respective normal counterparts (Fig. 3 C). On the other hand, the normal HPS-12 and cancer-associated HPS-13 stromal cells were found to have similar colony formation in two separate experiments. In two of the three matched pairs, cancer-associated prostate stromal clones appeared to be more resistant to serum starvation than the normal stromal cells.
Cancer-associated prostate stromal cells rescue LNCaP prostate cancer cells from starvation-induced death.
Stromal cells of the tumor microenvironment may promote cancer cell survival through mechanisms of cancer–stromal interaction (Sung and Chung 2002; Kogan-Sakin et al. 2009). Since the prostate stromal clones were insensitive to androgen and were highly resistant to serum starvation (Figs. 2 and 3), we assessed whether the stromal clones could promote survival of prostate cancer cells under similar conditions. The study was conducted by co-culturing the stromal cells with LNCaP prostate cancer cells. To prepare for co-culture, stromal cells were first grown to form a confluent monolayer completely covering the plastic surface of the culture ware. After removal of the culture medium, LNCaP prostate cancer cells were overlaid onto the monolayer in low density, so the co-culture was comprised of equal numbers of stromal and cancer cells.
Cells in the co-culture were treated with serum starvation for 60 d. During the treatment, most of the LNCaP cells died gradually, as seen by microscopic inspection and detected by markedly reduced PSA in the culture medium (Fig. 4 A). The low PSA level, however, was sustained to the end of the experiment at 60 d. Since none of the prostate stromal clones expressed detectable levels of PSA by ELISA (Fig. 4 A and data not shown), these results suggested that some LNCaP cells had survived the starvation.
Stromal cells render LNCaP cells resistant to starvation-induced death. Representative results from co-culture with the HPS-15 stromal clone are shown. A PSA production in the co-cultures (LNCaP+HPS-15 and RL-1+HPS-15) was detected throughout the serum starvation. In comparison, PSA was lost when cancer cells alone (LNCaP and RL-1) were subjected to starvation. PSA is not expressed in HPS-15 stromal cells (HPS-15). Data represent the mean of triplicate assays. Standard deviation for each data is less than 5% of the mean and is not shown. B When RL-1 cells were cultured alone, the loss of PSA production was due to cell death under serum starvation. Shown are cultures of RL-1 alone at the beginning (Day 1) and at Day 35 of serum starvation. All RL-1 cells died after 35 d of serum starvation. For fluorescence photography, a phase contrast microphotograph (upper panel) and red fluorescence image (lower panel) of the same field are shown. C Co-cultures of RL-1 with HPS-15 at the beginning (Day 1) and end (Day 60) of serum starvation. Representative surviving RL-1 cells (arrows) are shown among stromal cells. D After a 60-d serum starvation, surviving RL-1 cells from the co-culture formed colonies (arrows) among stromal cell colonies upon replating in normal culture medium for 14 d. Two representative results are shown.
To confirm the survival of LNCaP cells, we included in the experiment a red fluorescent LNCaP clone, RL-1, which stably expressed an AsRed2 red fluorescence protein. Similar to the parental LNCaP cells, the RL-1 clone was sensitive to serum starvation (Fig. 4 A and B). The co-cultures were subjected to serum starvation for 60 d, and survival of the RL-1 was documented by fluorescence microscopy. Although the number of red fluorescent cells became fewer as serum starvation progressed, these cells never disappeared (Fig. 4 C). Moreover, at the end of the experiment when the mixed cells in co-culture were replated in regular culture medium for colony formation, a substantial number of red fluorescent colonies were formed (Fig. 4 D). These results were in contrast to the control group, in which RL-1 cells alone were subjected to serum starvation. Cells in the control group all perished around 35 d (Fig. 4 B) and following the 60-d starvation, no red fluorescent colonies were formed in colony formation assays. This study indicated that by co-culture, prostate stromal cells could rescue LNCaP cells from starvation-induced death.
Compared to their normal counterparts, the cancer-associated HPS-11 and HPS-15 stromal cells displayed preferential protection to the LNCaP prostate cancer cells. In colony formation assays, when survival of the cancer cells was quantified following a 60-d serum starvation, 271.3 ± 57.1 red fluorescent colonies were formed from a triplicate co-culture of RL-1 with cancer-associated HPS-11 stromal cells, comparing to 61 ± 11 colonies from the co-cultures with the normal HPS-10 stromal cells (P < 0.05). Similarly, significantly more red fluorescent colonies were formed from the co-culture with cancer-associated HPS-15 stromal cells (613.3 ± 187.7) than with the normal HPS-14 stromal cells (81 ± 18.5). The results from these studies indicated that cancer-associated stromal cells showed stronger tropism on survival of the LNCaP prostate cancer cells.
Discussion
In this work, we established three matched pairs of normal and cancer-associated human prostate stromal cell lines. These cell cultures represent a rare collection of cells that can be used to explore cancer–stromal interaction. We describe the initial characterization of these cell lines at the morphologic and behavioral levels. By extracting common morphologic and behavioral features in three matched pairs, this study may provide a general view of the prostate stromal cells in culture.
This study demonstrates that prostate stromal cells are androgen-insensitive and are highly resistant to serum starvation (Figs. 2 and 3). This feature may be a reflection of the prostate stromal cells in clinical cases and animal studies, where androgen removal shows differentiated effects on epithelial and stromal cells (English et al. 1985; Sagalowsky 1985; Lippert and Keefer 1987; McConnell 1990; Sensibar et al. 1990). The cause of the androgen insensitivity and starvation resistance is poorly defined. Since the results were obtained from isolated stromal cells cultured in a defined medium, this study may suggest that androgen independence and starvation resistance are inherent properties of prostate stromal cells, sustained by endogenous mechanisms. The prostate stromal clones will be ideal subjects for investigating the molecular mechanisms underlying these properties.
We determined that cancer-associated prostate stromal cells have differential capabilities to survive serum starvation, as two of the three cancer-associated stromal clones formed significantly more colonies than the normal counterparts (Fig. 3 C). Moreover, the HPS-11 and HPS-15 cancer-associated stromal cells displayed increased potential to rescue LNCaP cells from serum starvation. These results are in agreement with the findings that cancer-associated stromal cells in the tumor microenvironment may have pathophysiologic behaviors (Ayala et al. 2003; Micke and Ostman 2004). We are currently performing comparative analyses on these matched stromal pairs in order to identify expressional changes supporting these behaviors.
We found few dramatic differences between the normal and cancer-associated prostate stromal cells as far as morphology and proliferation are concerned. The matched pairs showed mutually similar large shapes and slow growth (Figs. 1 and 2). On the other hand, both the cellular morphology and proliferation rate are quite distinct from those of the prostate cancer cells (Fig. 4 C and D). In the study of cancer–stromal interaction, the distinction may facilitate cell tracking during co-culture of cancer and stromal cells. Together with the fluorescently labeled LNCaP prostate cancer cells (Fig. 4 B), these stromal cells are valuable tools for in vitro investigation of cancer–stromal interaction in prostate cancer progression and metastasis.
Using in vitro co-culture to simulate cancer–stromal interaction, this study revealed that prostate stromal cells could rescue LNCaP prostate cancer cells from serum starvation (Fig. 4). Cultured alone, growth of the LNCaP cells was reduced by androgen deprivation, and all the LNCaP cells were killed by serum starvation (Fig. 4 B). In the presence of stromal cells, some cancer cells survived and maintained a latent state for an extended time (Fig. 4 C). Under favorable conditions, these cells grew to form colonies in large populations (Fig. 4 D). This finding may be clinically relevant because survival of cancer cells under deprivation conditions may cause tumor latency and cancer recurrence (Koivisto et al. 1996; Craft et al. 1999; Thalmann et al. 2000; Tso et al. 2000; Schroder 2008). It is likely that a key mechanism by which prostate stromal cells facilitate cancer progression is to sustain the viability of the cancer cells through cancer–stromal interaction. Further investigation into the cancer–stromal interaction under starvation conditions may unveil the molecular mechanism mediating the effect. The established cancer–stromal co-culture system described in this work is a simplified and reproducible experimental model for such studies.
Conclusions
In this study, we isolated and characterized three matched pairs of human prostate stromal clones. Compared to normal counterparts, two of the three cancer-associated stromal clones showed significantly higher resistance to serum starvation and stronger protection of LNCaP prostate cancer cells from starvation-induced death. The matched stromal cell pairs may serve as models for comparative analysis of molecular changes in the tumor microenvironment. These cells can also be used in co-culture with prostate cancer cells to simulate cancer–stromal interaction in the tumor microenvironment in order to define the role of prostate stromal cells in prostate cancer progression and metastasis.
References
Ayala G.; Tuxhorn J. A.; Wheeler T. M.; Frolov A.; Scardino P. T.; Ohori M.; Wheeler M.; Spitler J.; Rowley D. R. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin. Cancer Res. 9(13): 4792–4801; 2003.
Chung L. W. The role of stromal-epithelial interaction in normal and malignant growth. Cancer. Surv. 23: 33–42; 1995.
Chung L. W.; Cunha G. R. Stromal-epithelial interactions: II. Regulation of prostatic growth by embryonic urogenital sinus mesenchyme. Prostate. 4(5): 503–511; 1983.
Condon M. S. The role of the stromal microenvironment in prostate cancer. Semin. Cancer Biol. 15(2): 132–137; 2005.
Craft N.; Chhor C.; Tran C.; Belldegrun A.; DeKernion J.; Witte O. N.; Said J.; Reiter R. E.; Sawyers C. L. Evidence for clonal outgrowth of androgen-independent prostate cancer cells from androgen-dependent tumors through a two-step process. Cancer Res. 59(19): 5030–5036; 1999.
Cunha G. R. Role of mesenchymal-epithelial interactions in normal and abnormal development of the mammary gland and prostate. Cancer. 74(3 Suppl): 1030–1044; 1994.
Cunha G. R.; Cooke P. S.; Kurita T. Role of stromal-epithelial interactions in hormonal responses. Arch. Histol. Cytol. 67(5): 417–434; 2004.
Cunha G. R.; Foster B.; Thomson A.; Sugimura Y.; Tanji N.; Tsuji M.; Terada N.; Finch P. W.; Donjacour A. A. Growth factors as mediators of androgen action during the development of the male urogenital tract. World J. Urol. 13(5): 264–276; 1995.
Cunha G. R.; Hayward S. W.; Wang Y. Z. Role of stroma in carcinogenesis of the prostate. Differentiation. 70(9–10): 473–485; 2002.
English H. F.; Drago J. R.; Santen R. J. Cellular response to androgen depletion and repletion in the rat ventral prostate: autoradiography and morphometric analysis. Prostate. 7(1): 41–51; 1985.
Fasciana C.; van der Made A. C.; Faber P. W.; Trapman J. Androgen regulation of the rat keratinocyte growth factor (KGF/FGF7) promoter. Biochem. Biophys. Res. Commun. 220(3): 858–863; 1996.
Gleave M.; Hsieh J. T.; Gao C. A.; von Eschenbach A. C.; Chung L. W. Acceleration of human prostate cancer growth in vivo by factors produced by prostate and bone fibroblasts. Cancer Res. 51(14): 3753–3761; 1991.
Harding M. A.; Theodorescu D. Prostate tumor progression and prognosis. interplay of tumor and host factors. Urol. Oncol. 5(6): 258–264; 2000.
He H.; Yang X.; Davidson A. J.; Wu D.; Marshall F. F.; Chung L. W.; Zhau H. E.; Wang R. Progressive epithelial to mesenchymal transitions in ARCaPE prostate cancer cells during xenograft tumor formation and metastasis. Prostate 70: 518–528; 2009.
Kogan-Sakin I.; Cohen M.; Paland N.; Madar S.; Solomon H.; Molchadsky A.; Brosh R.; Buganim Y.; Goldfinger N.; Klocker H.; Schalken J. A.; Rotter V. Prostate stromal cells produce CXCL-1, CXCL-2, CXCL-3 and IL-8 in response to epithelia-secreted IL-1. Carcinogenesis. 30(4): 698–705; 2009.
Koivisto P.; Visakorpi T.; Kallioniemi O. P. Androgen receptor gene amplification: a novel molecular mechanism for endocrine therapy resistance in human prostate cancer. Scand. J. Clin. Lab. Invest. Suppl. 226: 57–63; 1996.
Lippert M. C.; Keefer D. A. Prostate adenocarcinoma: effects of castration on in situ androgen uptake by individual cell types. J. Urol. 137(1): 140–145; 1987.
Loberg R. D.; Gayed B. A.; Olson K. B.; Pienta K. J. A paradigm for the treatment of prostate cancer bone metastases based on an understanding of tumor cell-microenvironment interactions. J. Cell. Biochem. 96(3): 439–446; 2005.
Lu W.; Luo Y.; Kan M.; McKeehan W. L. Fibroblast growth factor-10. A second candidate stromal to epithelial cell andromedin in prostate. J. Biol. Chem. 274(18): 12827–12834; 1999.
McConnell J. D. Androgen ablation and blockade in the treatment of benign prostatic hyperplasia. Urol. Clin. North Am. 17(3): 661–670; 1990.
Micke P.; Ostman A. Tumour-stroma interaction: cancer-associated fibroblasts as novel targets in anti-cancer therapy? Lung Cancer. 45(Suppl 2): S163–S175; 2004.
Planz B.; Aretz H. T.; Wang Q.; Tabatabaei S.; Kirley S. D.; Lin C. W.; McDougal W. S. Immunolocalization of the keratinocyte growth factor in benign and neoplastic human prostate and its relation to androgen receptor. Prostate. 41(4): 233–242; 1999.
Rhee H. W.; Zhau H. E.; Pathak S.; Multani A. S.; Pennanen S.; Visakorpi T.; Chung L. W. Permanent phenotypic and genotypic changes of prostate cancer cells cultured in a three-dimensional rotating-wall vessel. In Vitro Cell Dev. Biol. Anim. 37(3): 127–140; 2001.
Sagalowsky A. I. Endocrine therapy for prostate cancer. Spec. Top Endocrinol. Metab. 7: 101–129; 1985.
Schroder F. H. Progress in understanding androgen-independent prostate cancer (AIPC): a review of potential endocrine-mediated mechanisms. Eur. Urol. 53(6): 1129–1137; 2008.
Sensibar J. A.; Liu X. X.; Patai B.; Alger B.; Lee C. Characterization of castration-induced cell death in the rat prostate by immunohistochemical localization of cathepsin D. Prostate. 16(3): 263–276; 1990.
Singh G.; Lakkis C. L.; Laucirica R.; Epner D. E. Regulation of prostate cancer cell division by glucose. J. Cell Physiol. 180(3): 431–438; 1999.
Sung S. Y.; Chung L. W. Prostate tumor-stroma interaction: molecular mechanisms and opportunities for therapeutic targeting. Differentiation. 70(9–10): 506–521; 2002.
Thalmann G. N.; Sikes R. A.; Wu T. T.; Degeorges A.; Chang S. M.; Ozen M.; Pathak S.; Chung L. W. LNCaP progression model of human prostate cancer: androgen-independence and osseous metastasis. Prostate. 44(2): 91–103; 2000.
Tso C. L.; McBride W. H.; Sun J.; Patel B.; Tsui K. H.; Paik S. H.; Gitlitz B.; Caliliw R.; van Ophoven A.; Wu L.; deKernion J.; Belldegrun A. Androgen deprivation induces selective outgrowth of aggressive hormone-refractory prostate cancer clones expressing distinct cellular and molecular properties not present in parental androgen-dependent cancer cells. Cancer J. 6(4): 220–233; 2000.
Wu H. C.; Hsieh J. T.; Gleave M. E.; Brown N. M.; Pathak S.; Chung L. W. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int. J. Cancer. 57(3): 406–412; 1994.
Xu J.; Wang R.; Xie Z. H.; Odero-Marah V.; Pathak S.; Multani A.; Chung L. W.; Zhau H. E. Prostate cancer metastasis: role of the host microenvironment in promoting epithelial to mesenchymal transition and increased bone and adrenal gland metastasis. Prostate. 66(15): 1664–1673; 2006.
Yan G.; Fukabori Y.; Nikolaropoulos S.; Wang F.; McKeehan W. L. Heparin-binding keratinocyte growth factor is a candidate stromal-to-epithelial-cell andromedin. Mol. Endocrinol. 6(12): 2123–2128; 1992.
Acknowledgments
This work is supported by research grants R21CA112330, PC040578, CA132388 (RXW), and CA98912-02 (LWKC).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: J. Denry Sato
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sun, X., He, H., Xie, Z. et al. Matched pairs of human prostate stromal cells display differential tropic effects on LNCaP prostate cancer cells. In Vitro Cell.Dev.Biol.-Animal 46, 538–546 (2010). https://doi.org/10.1007/s11626-010-9309-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-010-9309-z