Summary
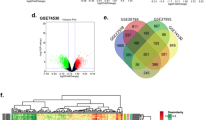
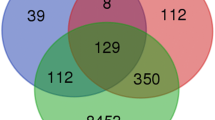
Human tongue cancer (TC) is an aggressive malignancy with a very poor prognosis. There is an urgent need to elucidate the underlying molecular mechanisms involved in TC progression. mRNA expression profiles play a vital role in the exploration of cancer-related genes. Therefore, the purpose of our study was to identify the progression associated candidate genes of TC by bioinformatics analysis. Five microarray datasets of TC samples were downloaded from the Gene Expression Omnibus (GEO) database and the data of 133 TC patients were screened from The Cancer Genome Atlas (TCGA) head and neck squamous cell carcinoma (HNSC) database. The integrated analysis of five microarray datasets and the RNA sequencing data of TC samples in TCGA-HNSC was performed to obtain 1023 overlapping differentially expressed genes (DEGs) in TC and adjacent normal tissue (ANT) samples. Next, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted to enrich the significant pathways of the 1023 DEGs and PI3KAkt signaling pathway (P=0.011) was selected to be the candidate pathway. A total of 23 DEGs with |log2 fold change (FC)| ≥1.0 in phosphatidylinositol 3-kinase-serine/threonine kinase (PI3K-Akt) signaling pathway were subjected to survival analysis of 125 eligible TC samples in TCGA database, indicating increased integrin-α3 gene (ITGA3) expression was significantly associated with poorer prognosis. Taken together, our study suggested ITGA3 may facilitate the development of TC via activating PI3K-Akt signaling pathway.
Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin, 2016, 66(1):7–30
Kokemueller H, Rana M, Rublack J, et al The Hannover experience: Suigical treatment of tongue cancer--A clinical retrospective evaluation over a 30 years period. Head Neck Oncol, 2011, 3(1):27
Dong Y, Zhao Q, Ma X, et al. Establishment of a new OSCC cell line derived from OLK and identification of malignant transformation-related proteins by differtial proteomics approach. Sci Rep, 2015, 5:12 668
Broadbent J, Sampson D, Sabapathy S, et al. Gene networks in skeletal muscle following endurance exercise are coexpressed in blood neutrophils and linked with blood inflammation markers. J Appl Physiol, 2017, 122(4):752–766
Li G, Li X, Yang M, et al. Prediction of biomarkers of oral squamous cell carcinoma using microarray technology. Sci Rep, 2017, 7:42 105
Randhawa V, Achaiya V. Integrated network analysis and logistic regression modeling identify stagespecific genes in oral squamous cell carcinoma. BMC Med Genomics, 2015, 8(1):39
Kang DD, Sibille E, Kaminski N, et al MetaQC: objective quality control and inclusion/exclusion criteria for genomic meta-analysis. Nucleic Acids Res, 2011, 40(2):el5–el5
Wang X, Kang DD, Sh i K, et al. Ail R package suite for microarray meta-analysis in quality control, differentially expressed gene analysis and pathway enrichment detection. Bioinformatics, 2012, 28(19):2534–2536
Qi C, Hong L, Cheng Z, et al. Identification of metastasis associated genes in colorectal cancer using metaDE and survival analysis. Oncol Lett, 2016, 11:568–574
Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations far microarray meta-analysis. Nucleic Acids Res, 2012, 40(9):3785–3799
Sun M, Sun T, He Z, et ál. Identification of two novel biomarkers of rectal carcinoma progression and prognosis via co-expression network analysis. Qncotarget, 2017, 8(41):69 594
Yu G, Wang L G, Han Y, et al clusterProfiler: an R package for compaiing biological themes among gene clusters. OMICS, 2012, 16(5):284–287
Lo WY, Tsai MH, Tsai Y, et al. Identification of overexpressed proteins in oral squamous cell carcinoma (OSCC) patients by clinical proteomic analysis. Clin Chim Acta, 2007, 376(1–2):101–107
Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol, 2011, 3(3):a004994
Bendas G, Borsig L. Cancer cell adhesion and metastasis: selectins, integrins, and the inhibitory potential of heparins. Int J Cell Biol, 2012, 2012:676 731
Koshizuka K, Hanazawa T, Kikkawa N, et al. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci, 2017, 108(8):1681–1692
Tang XR, Wen X, He QM, et al. MicroRNA-101 inhibits invasion and angiogenesis through targeting ITGA3 and its systemic delivery inhibits lung metastasis in nasopharyngeal carcinoma. Cell Death Dis, 2017, 8(1):e2566
Sakaguchi T, Yoshino H, Yonemori M, et al. Regulation of ITGA3 by the dual-stranded microRNA-199 family as a potential prognostic marker in bladder cancer. Br J Cancer, 2017, 116(8):1077
Linhares MM, Affonso Jr RJ, de Souza Viana L, et al. Genetic and immunohistochemical expression of integrins ITGAV, ITGA6, and ITGA3 as prognostic factor for colorectal cancer: models for global and disease-free survival. PLoS One, 2015, 10(12):e0144 333
Chen J, Xu X, Wang H. Expression of integrin-α 3 mRNA in meningiomas and its correlation with proliferation and invasion. J Huazhong Univ Sci Technolog Med Sci, 2009, 29(1):94–96
Author information
Authors and Affiliations
Corresponding authors
Additional information
This project was supported by grants from Natural Science Foundation of Hubei Province (No. 220100321) and Clinical Medical Research Center of Peritoneal Cancer of Wuhan (No. 2015060911020462).
Rights and permissions
About this article
Cite this article
Chen, Wl., Wang, Xk. & Wu, W. Identification of ITGA3 as an Oncogene in Human Tongue Cancer via Integrated Bioinformatics Analysis. CURR MED SCI 38, 714–720 (2018). https://doi.org/10.1007/s11596-018-1935-9
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-018-1935-9