Summary
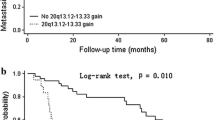
Copy number aberrations (CNAs) in chromosome arm 8q have been associated with unfavorable clinical outcomes of several cancers and progressive tumor characteristics of hepatocellular carcinoma (HCC). This study was to identify correlation of CNAs in 8q with clinical outcomes of HCC patients, and further screen for differentially expressed genes in outcome-related CNAs. Array comparative genomic hybridization and expression arrays were performed to detect CNAs and expression levels, respectively. The correlations between CNAs in 8q and outcomes were analyzed in 66 patients, with a median follow-up time of 45.0 months (range, 2.6-108.6 months). One hundred and nine cases were further evaluated to identify differentially expressed genes in the potential outcome-related CNAs. Copy number gain in 8q was observed in 22 (33.3%) of the 66 HCC cases. The most recurrent gains (with frequencies >20%) were 8q13.3-21.3,8q21.3-23.3,8q23.3-24.13,8q24.13-24.3, and 8q24.3. Survival analysis showed that 8q24.13-24.3 gain was significantly associated with reduced overall survival (jP=0.010). Multivariate Cox analysis identified 8q24.13-24.3 gain as an independent prognostic factor for poor overall survival (HR=2.47; 95% CI=1.16-5.26; Р=0.019). Apanel of 17 genes within the 8q24.13-24.3 region, including ATAD2,SQLE,PVT1,ASAP1, and NDRG1 were significantly upregulated in HCCs with 8q24.13-24.3 gain compared to those without. These results suggest that copy number gain at 8q24.13-24.3 is an unfavorable prognostic marker for HCC patients, and the potential oncogenes ATAD2,SQLE, PVT1, ASAP1,and NDRG1 within the regional gain, may contribute coordinately to the 8q24.13-24.3 gain-related poor prognosis.
Similar content being viewed by others
References
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. С A Cancer J Clin, 2016, 66(2):115–132
Pinato DJ, Pirisi M, Maslen L, et al. Tissue biomarkers of prognostic significance in hepatocellular carcinoma. Adv Anat Pathol, 2014, 21(4):270–284
Shlien A, Maikin D. Copy number variations and cancer susceptibility. Curr Opin Oncol, 2010, 22(1):55–63
Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature, 2010, 463(7283):899–905
Walker LC, McDonald M, Wells JE, et al. Dualcolor fluorescence in situ hybridization reveals an association of chromosome 8q22 but not 8p21 imbalance with high grade invasive breast carcinoma. PLoS One, 2013, 8(7):e70790
Cheng I, Levin AM, Tai YC, et al. Copy number alterations in prostate tumors and disease aggressiveness. Genes Chromosomes Cancer, 2012, 51(1)r66–76
Vékony H, Roser K, Loning T, et al. Copy number gain at 8q12.1–q22.1 is associated with a malignant tumor phenotype in salivary gland myoepitheliomas. Genes Chromosomes Cancer, 2009, 48(2):202–212
Silva MP, Barros-Silva JD, Ersvaer E, et al. Cancer Prognosis Defined by the Combined Analysis of 8q, PTEN and ERG. Transi Oncol, 2016, 9(6):575–582
Zafarana G, Ishkanian AS, Malloff CA, et al. Copy number alterations of c-MYC and PTEN are prognostic factors for relapse after prostate cancer radiotherapy. Cancer, 2012, 118(16):4053–4062
Yoshioka S, Tsukamoto Y, Hijiya N, et al. Genomic profiling of oral squamous cell carcinoma by arraybased comparative genomic hybridization. PLoS One, 2013, 8(2):e56165
Munoz-Bellvis L, Fontanillo C, Gonzalez-Gonzalez M, et al. Unique genetic profile of sporadic colorectal cancer liver metastasis versus primary tumors as defined by high-density single-nucleotide p ymorphism arrays. Mod Pathol, 2012, 25(4):590–601
Boelens MC, Kok K, van der Vlies P, et al. Genomic aberrations in squamous cell lung carcinoma related to lymph node or distant metastasis. Lung Cancer, 2009, 66(3):372–378
Vincent-Chong VK, Salahshourifar I, Woo KM, et al. Genome wide profiling in oral squamous cell carcinoma identifies a four genetic marker signature of prognostic significance. PLoS One, 2017, 12(4):e0174865
Klatte T, Kroeger N, Rampersaud EN, et al. Gain of chromosome 8q is associated with metastases and poor survival of patients with clear cell renal cell carcinoma. Cancer, 2012, 118(23):5777–5782
El Gammal AT, Bruchmann M, Zustin J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res, 2010, 16(1):56–64
Lagerstedt KK, Staaf J, Jonsson G, et al. Tumor genome wide DNA alterations assessed by array CGH in patients with poor and excellent survival following operation for colorectal cancer. Cancer Inform, 2007, 3:341–355
Schleicher C, Poremba C, Wolters H, et al. Gain of chromosome 8q: a potential prognostic marker in resectable adenocarcinoma of the pancreas? Ann Surg Oncol, 2007, 14(4):1327–1335
Milioli HH, Tishchenko I, Riveros C, et al. Basal-like breast cancer: molecular profiles, clinical features and survival outcomes. BMC Med Genomics, 2017, 10(1):19
Okamoto H, Yasui K, Zhao C, et al PTK2 and EIF3 S3 genes may be amplification targets at 8q23–q24 and are associated with large hepatocellular carcinomas. Hepatology, 2003, 38(5):1242–1249
Chochi Y, Kawauchi S, Nakao M, et al. A copy number gain of the 6p arm is linked with advanced hepatocellular carcinoma: an array-based comparative genomic hybridization study. J Pathol, 2009, 217(5):677–684
Liu YJ, Zhou Y, Yeh MM. Recurrent genetic alterations in hepatitis C-associated hepatocellular carcinoma detected by genomic microarray: a genetic, clinical and pathological correlation study. Mol Cytogenet, 2014, 7(1):81
Versluis M, de Lange MJ, van Pelt SI, et al. Digital PCR validates 8q dosage as prognostic tool in uveal melanoma. PLoS One, 2015, 10(3):e0116371
Han S, Park K, Shin E, et al. Genomic change of chromosome 8 predicts the response to taxane-based neoadjuvant chemotherapy in node-positive breast cancer. Oncol Rep, 2010, 24(1):121–128
Hwang HW, Ha SY, Bang H, et al ATAD2 as a Poor Prognostic Marker for Hepatocellular Carcinoma after Curative Resection. Cancer Res Treat, 2015, 47(4):853–861
Wu G, Lu X, Wang Y, et al. Epigenetic high regulation of ATAD2 regulates the Hh pathway in human hepatocellular carcinoma. Int J Oncol, 2014, 45(1):351–361
Brown DN, Caffa I, Cirmena G, et al. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sei Rep, 2016, 6:19435
Zhang HY, Li HM, Yu Z, et al. Expression and significance of squalene epoxidase in squamous lung cancerous tissues and pericarcinoma tissues. Thorac Cancer, 2014, 5(4):275–280
Colombo T, Farina L, Macino G, et al. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int, 2015, 2015:304208
Ding C, Yang Z, Lv Z, et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett, 2015, 9(2):955–963
Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVTl as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett, 2017, 388:208–219
Song J, Wu X, Liu F, et al. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem Biophys Res Commun, 2017, 490(2):217–224
Huang C, Liu S, Wang H, et al LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transi Res, 2016, 8(ll):5025–5034
Muller T, Stein U, Poletti A, et al ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene, 2010, 29(16):2393–2403
Li M, Tian L, Yao H et al. ASAP1 mediates the invasive phenotype of human laryngeal squamous cell carcinoma to affect survival prognosis. Oncol Rep, 2014, 31(6):2676–2682
Hou T, Yang C, Tong C, et al. Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. Int J Clin Exp Pathol, 2014, 7(1):280–287
Song Y, Cao L. N-myc downstream-regulated gene 1: Diverse and complicated functions in human hepatocellular carcinoma (Review). Oncol Lett, 2013, 6(6):1539–1542
Cheng J, Xie HY, Xu X, et al NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer Lett, 2011, 310(1):35–45
Shen JY, Li C, Wen TF, et al. Alpha fetoprotein changes predict hepatocellular carcinoma survival beyond the Milan criteria after hepatectomy. J Surg Res, 2017, 209:102–111
Sun L, Liu T, Zhang S, et al. Oct4 induces EMT through LEF1/beta-catenin dependent WNT signaling pathway in hepatocellular carcinoma. Oncol Lett, 2017, 13(4):2599–2606
Katkoori VR, Shanmugam C, Jia X, et al. Prognostic significance and gene expression profiles of p53 mutations in microsatellite-stable stage III colorectal adenocarcinomas. PLoS One, 2012, 7(1):e30020
Author information
Authors and Affiliations
Corresponding authors
Additional information
This project was supported by grants from the Medical Science and Technology Innovation Fund of PLA, Nanjing branch, China (No. 14ZD07; 08MA023) and Ningbo Nature Science Foundation Program (No. 2009A610126).
Rights and permissions
About this article
Cite this article
Zhao, K., Zhao, Y., Zhu, Jy. et al. A Panel of Genes Identified as Targets for 8q24.13-24.3 Gain Contributing to Unfavorable Overall Survival in Patients with Hepatocellular Carcinoma. CURR MED SCI 38, 590–596 (2018). https://doi.org/10.1007/s11596-018-1918-x
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-018-1918-x