Abstract
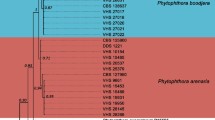
A new species of Phytophthora was isolated from stem and root rot of chrysanthemum in the Gifu and Toyama prefectures of Japan. The species differs from other Phytophthora species morphologically, and is characterized by nonpapillate, noncaducous sporangia with internal proliferation, formation of both hyphal swellings and chlamydospores, homothallic nature, distinctive intercalary antheridia, and funnel-shaped oogonia. The new species can grow even at 35°C, with an optimum growth temperature of 30°C in V8 juice agar medium. In phylogenetic analyses based on five nuclear regions (LSU rDNA; genes for translation elongation factor 1α, β-tubulin, 60 S ribosomal protein L10, and heat shock protein 90), the isolates formed a monophyletic clade. Although the rDNA ITS region shows a high resolution and has proven particularly useful for the separation of Phytophthora species, it was difficult to align the sequences for phylogenetic analysis. Therefore, ITS region analysis using related species as defined by the multigene phylogeny was performed, and the topology of the resulting tree also revealed a monophyletic clade formed by the isolates of the species. The morphological characteristics and phylogenetic relationships indicate that the isolates represent a new species, Phytophthora chrysanthemi sp. nov. In pathogenicity tests, chrysanthemum plants inoculated with the isolates developed lesions on stems and roots within 3 days, and the symptoms resembled the ones originally observed. Finally, the pathogen’s identity was confirmed by re-isolation from lesions of infected plants.






Similar content being viewed by others
References
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Ann PJ, Ko WH (1980) Phytophthora insolita, a new species from Taiwan. Mycologia 72:1180–1185
Beakes GW, Sekimoto S (2009) The evolutionary phylogeny of oomycetes-insights gained from studies of holocarpic parasites of algae and invertebrates. In: Lamour K, Kamoun S (eds) Oomycete genetics and genomics: diversity, interactions and research tools. Wiley, New York, pp 1–23
Belbahri L, Moralejo E, Calmin G, Oszako T, Garcia JA, Descals E, Lefort F (2006) Phytophthora polonica, a new species isolated from declining Alnus glutinosa stands in Poland. FEMS Microbiol Lett 261:165–174
Blair JE, Coffey MD, Park SY, Geiser DM, Kang S (2008) A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Gen Biol 45:266–277
Brasier CM, Beales PA, Kirk SA, Denman S, Rose J (2005) Phytophthora kernoviae sp. nov., an invasive pathogen causing bleeding stem lesions on forest trees and foliar necrosis of ornamentals in the UK. Mycol Res 109:853–859
Bremer K (1988) The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42:795–803
Bremer K (1994) Branch support and tree stability. Cladistics 10:295–304
Bruns TD, White TJ, Taylor JW (1991) Fungal molecular systematic. Annu Rev Ecol Syst 22:525–564
Chikuo Y, Morikawa T, Kageyama K (2007) Addition of the Pathogen of Phytophthora Root Rot of Chrysanthemum (Chrysanthemum morifolium). Annu Phytopathol Soc Jpn 73:180
Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM (2000) A molecular phylogeny of Phytophthora and related oomycetes. Fungal Gen Biol 30:17–32
Dick MA, Dobbie K, Cooke DEL, Brasier CM (2006) Phytophthora captiosa sp. nov. and P. fallax sp. nov. causing crown dieback of Eucalyptus in New Zealand. Mycol Res 110:393–404
Donahoo R, Blomquist CL, Thomas SL, Moulton JK, Cooke DEL, Lamour KH (2006) Phytophthora foliorum sp. nov., a new species causing leaf blight of azalea. Mycol Res 110:1309–1322
Donoghue MJ, Olmstead RG, Smith JF, Palmer JD (1992) Phylogenetic relationships of dipsacales based on rbcL sequences. Ann Mo Bot Gard 79:333–345
Erwin DC, Ribeiro OK (1996) Phytophthora diseases worldwide. American Phytopathological Society, St Paul, Minnesota
Felsenstein J (1985) Confidence limits on phylog-enies: an approach using the bootstrap. Evolution 39:783–791
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Ivors KL, Bonants PJM, Rizzo DM, Garbelotto M (2004) AFLP and phylogenetic analyses of North American and European populations of Phytophthora ramorum. Mycol Res 108:378–392
Jobb G, von Haeseler A, Strimmer K (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4:18
Kageyama K, Senda M, Asano T, Suga H, Ishiguro K (2007) Intra-isolate heterogeneity of the ITS region of rDNA in Pythium helicoides. Mycol Res 111:416–423
Kagiwata T (1972) A Phytophthora blight of chrysanthemum caused by Phytophthora cactorum. Nougaku shuho 30:176–185 (In Japanese)
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29:170–179
Ko WH, Ann PJ (1985) Phytophthora humicola, a new species from soil of a citrus orchard in Taiwan. Mycologia 77:631–636
Kroon LPNM, Bakker FT, van den Bosch GBM, Bonants PJM, Flier WG (2004) Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet Biol 41:766–782
Lee SB, Taylor JW (1992) Phylogeny of five fungus-like Protoctistan Phytophthora species, inferred from the Internal Transcribed Spacers of Ribosomal DNA. Mol Biol Evol 9:636–653
Martin FN, Tooley PW (2003a) Phylogenetic relationships among Phytophthora species inferred from sequence analysis of the mitochondrially-encoded cytochrome oxidase I and II genes. Mycologia 95:269–284
Martin FN, Tooley PW (2003b) Phylogenetic relationships of Phytophthora ramorum, P. nemorosa, and P. pseudosyringae, three species recovered from areas in California with sudden oak death. Mycol Res 107:1379–1391
Masago H, Yoshikawa M, Fukuda M, Nakanishi N (1977) Selective inhibition of Pythium spp. on a medium for direct isolation of Phytophthora spp. from soil and plants. Phytopathology 67:425–428
Matsumoto C, Kageyama K, Suga H, Hyakumachi M (2000) Intraspecific DNA polymorphisms of Pythium irregulare. Mycol Res 104:1333–1341
Mostowfizadeh-Ghalamfarsa R, Cooke DEL, Banihasemi Z (2008) Phytophthora parsiana sp. nov., a new high-temperature tolerant species. Mycol Res 112:783–794
O’Donnell K (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp 225–233
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461–464
Sugiura N (1978) Further analysis of the data by Akaike’s information criterion and the finite corrections. Comm Stat Theory Methods 7:13–26
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony and other methods, 4.0β10. Sinauer, Sunderland
Tanabe AS (2007) Kakusan: a computer program to automate the selection of a nucleotide substitution model and the configuration of a mixed model on multilocus data. Mol Ecol Notes 7:962–964
Tsukiboshi T, Chikuo Y, Ito Y, Matsushita Y, Kageyama K (2007) Root and stem rot of chrysanthemum caused by five Pythium species in Japan. J Gen Plant Pathol 73:293–296
Villa NO, Kageyama K, Asano T, Suga H (2006) Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and beta-tubulin sequences. Mycologia 98:410–422
Vos RA (2003) Accelerated likelihood surface exploration: the likelihood ratchet. Syst Biol 52:368–373
Watanabe H, Horinouchi H, Ichihara S, Kuwabara K, Watanabe T, Kageyama K (2007) Occurrence of Phytophthora rot of chrysanthemum caused by Phytophthora sp. Annu Phytopathol Soc Jpn 73:60
Waterhouse GM (1963) Key to the species of Phytophthora de Bary. Mycol Pap No. 92
Waterhouse GE (1967) Key to Pythium Pringsheim. Commonw Mycol Inst Mycol Pap 109:1–15
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocol: a guide of methods and application. Academic, California, pp 315–322
Acknowledgments
We thank Dr. Susan A. Kirk (Forest Research Agency, Farnham), Dr. Lassaad Belbahri (Laboratory of Applied Genetics, Switzerland), Dr. Margaret Dick (New Zealand Forest Research Institute, New Zealand), Dr. Michael David Coffey (University of California Riverside, United States of America), and Dr. Reza Mostowfizadeh Ghalamfarsa (Department of Plant Protection, Siraz University, Iran) for providing isolates.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 159 kb)
Rights and permissions
About this article
Cite this article
Naher, M., Motohash, K., Watanabe, H. et al. Phytophthora chrysanthemi sp. nov., a new species causing root rot of chrysanthemum in Japan. Mycol Progress 10, 21–31 (2011). https://doi.org/10.1007/s11557-010-0670-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-010-0670-9