Abstract
Purpose
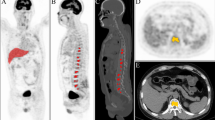
Multiple myeloma (MM) is a bone marrow cancer that accounts for 10% of all hematological malignancies. It has been reported that FDG PET imaging provides prognostic information for both baseline and therapeutic follow-up of MM patients using visual analysis. In this study, we aim to develop a computer-assisted method based on PET quantitative image features to assist diagnoses and treatment decisions for MM patients.
Methods
Our proposed model relies on a two-stage method with Random Survival Forest (RFS) and variable importance (VIMP) for both feature selection and prediction. The targeted variable for prediction is the progression-free survival (PFS). We consider texture-based (radiomics), conventional (e.g., SUVmax) and clinical biomarkers. We evaluate PFS predictions in terms of C-index and final prognosis separation in two risk groups, from a database of 66 patients who were part of the prospective multi-centric french IMAJEM study.
Results
Our method (VIMP + RSF) provides better results (1-C-index of 0.36) than conventional methods such as Lasso–Cox and gradient-boosting Cox (0.48 and 0.56, respectively). We experimentally proved the interest of using selection (0.61 for RSF without selection) and showed that VIMP selection is more stable and gives better results than minimal depth and variable hunting (0.47 and 0.43). The approach gives better prognosis group separation (a p value of 0.05 against 0.11 to 0.4 for others).
Conclusion
Our results confirm the predictive value of radiomics for MM patients, in particular, they demonstrate that quantitative/heterogeneity image-based features reduce the error of the predicted progression. To our knowledge, this is the first work using RFS on PET images for the progression prediction of MM patients. Moreover, we provide an analysis of the feature selection process, which points toward the identification of clinically relevant biomarkers.






Similar content being viewed by others
Notes
Right censoring occurs when no event (death/progression) has taken place at the end of the evaluation period.
The concordance index is the frequency of concordant pairs among all pairs of subjects.
References
Amin SB, Minvielle S, Hanlon B, Shah PK, Li C, Li Y, Swanson D, Moreau P, Magrangeas F, Anderson KC, Avet-Loiseau H, Munshi NC (2014) Gene expression profile alone is inadequate in predicting complete response in multiple myeloma. Leukemia 28:2229–2234
Breiman L (1996) Bagging predictors. Mach Learn 42(2):123–140
Bühnemann C, Li S, Yu H, White HB, Schäfer K, Llombart-Bosch A, Machado I, Picci P, Hogendoorn P, Athanasou N, Noble J, Hassa A (2014) Quantification of the heterogeneity of prognostic cellular biomarkers in ewing sarcoma using automated image and random survival forest analysis. Plos ONE 9(9):e107105
Carlier T, Bailly C, Leforestier R, Touzeau C, Moreau P, Bodere F, Bodet-Milin C (2017) Prognostic added value of PET textural features at diagnosis in multiple myeloma. J Nuclear Med 58(supplement 1):111
Cox DR (1972) Regression models and life-tables. J R Stat Soc Ser B (Methodol) 34(2):187–220
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images are more than pictures, they are data. Radiology 278:563–577
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC–3(6):610–621
Ishwaran H, Kogalur U, Blackstone E, Lauer M (2018) Random surival forest. Ann Appl Stat 2(3):841–860
Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS (2010) High-dimensional variable selection for survival data. J Am Stat Assoc 105(489):205–217
Kaplan EL, Meier P (1958) Nonparametric estimation from incomplete observations. J Am Stat Assoc 53(282):457–481
Lartizien C, Rogez M, Niaf E, Ricard F (2014) Computer-aided staging of lymphoma patients with FDG PET/CT imaging based on textural information. IEEE J Biomed Health Inform 18(3):946–955
McDonald JE, Kessler MM, Gardner MW, Buros AF, Ntambi JA, Waheed S, van Rhee F, Zangari M, Heuck CJ, Petty N, Schinke C, Thanendrarajan S, Mitchell A, Hoering A, Barlogie B, Morgan GJ, Davies FE (2017) Assessment of total lesion glycolysis by \(^{18}\)F FDG PET/CT significantly improves prognostic value of GEP and ISS in myeloma. Clin Cancer Res 23(8):1981–1987
Larue RTHM, Defraene G, Ruysscher DKMD, Lambin P, van Elmpt WJC (2017) Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. The Br J Radiol 90(1070):20160665
Moreau P, Caillon F, Bodet-Milin C (2017) Prospective evaluation of magnetic resonance imaging and [18F]Fluorodeoxyglucose positron emission tomography-computed tomography at diagnosis and before maintenance therapy in symptomatic patients with multiple myeloma included in the IFM/DFCI 2009 trial: Results of the IMAJEM study. J Clin Oncol 35(25):2911–2918
Ridgeway G (1999) The state of boosting. Comput Sci Stat. citeulike-article-id:7678637
Ridgeway G (2007) Generalized boosted models : a guide to the GBM package
Tibshirani R (1997) The lasso method for variable selection in the cox model. Stat Med 16(4):385–395
Vallières M, Kay-Rivest E, Perrin LJ, Liem X, Furstoss C, Aerts HJWL, Khaouam N, Nguyen-Tan PF, Wang CS, Sultanem K, Seuntjens J, Naqa IE (2017) Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep 7(10117):2911–2918
Wenzheng S, Jiang M, Dang J, Chang P, Yin FF (2018) Effect of machine learning methods on predicting NSCLC overall survival time based on Radiomics analysis. Radiat Oncol 13(1):197
Xu L, Tetteh G, Lipkova J, Zhao Y, Li H, Christ P, Piraud M, Buck A, Shi K, Menze BH (2018) Automated whole-body bone lesion detection for multiple myeloma on 68Ga-Pentixafor PET/CT imaging using deep learning methods. Contrast Media Mol Imaging 2018, Article ID 2391925. https://doi.org/10.1155/2018/2391925
Zamagni E, Patriarca F, Nanni C, Zannetti B, Englaro E, Pezzi A, Tacchetti P, Buttignol S, Perrone G, Brioli A, Pantani L, Terragna C, Carobolante F, Baccarani M, Fanin R, Fanti S, Cavo M (2011) Prognostic relevance of 18-F FDG PET/CT in newly diagnosed multiple myeloma patients treated with up-front autologous transplantation. Blood 118(23):5989–5995
Zhou Y, Mcardle JJ (2015) Rationale and applications of survival tree and survival ensemble methods. Psychometrika 80(3):811–33
Zwanenburg A, Leger S, Vallières M, Löck S (2016) Image biomarker standardisation initiative. arXiv:1612.07003
Funding
This work has been supported in part by the European Regional Development Fund, the Pays de la Loire region on the Connect Talent scheme MILCOM (Multi-modal Imaging and Learning for Computational-based Medicine), Nantes Métropole (Convention 2017-10470), the French National Agency for Research called “Investissements d’Avenir” IRON Labex \(\hbox {n}^{\mathrm{o}}\) ANR-11-LABX-0018-01 and INCa-DGOS-Inserm_12558 (SIRIC ILIAD)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Morvan, L., Carlier, T., Jamet, B. et al. Leveraging RSF and PET images for prognosis of multiple myeloma at diagnosis. Int J CARS 15, 129–139 (2020). https://doi.org/10.1007/s11548-019-02015-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-02015-y