Abstract
Purpose
Diagnosis and characterization of brain neoplasms appears of utmost importance for therapeutic management. The emerging of imaging techniques, such as Magnetic Resonance (MR) imaging, gives insight into pathology, while the combination of several sequences from conventional and advanced protocols (such as perfusion imaging) increases the diagnostic information. To optimally combine the multiple sources and summarize the information into a distinctive set of variables however remains difficult. The purpose of this study is to investigate machine learning algorithms that automatically identify the relevant attributes and are optimal for brain tumor differentiation.
Methods
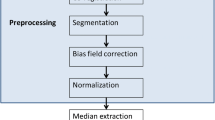
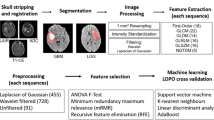
Different machine learning techniques are studied for brain tumor classification based on attributes extracted from conventional and perfusion MRI. The attributes, calculated from neoplastic, necrotic, and edematous regions of interest, include shape and intensity characteristics. Attributes subset selection is performed aiming to remove redundant attributes using two filtering methods and a wrapper approach, in combination with three different search algorithms (Best First, Greedy Stepwise and Scatter). The classification frameworks are implemented using the WEKA software.
Results
The highest average classification accuracy assessed by leave-one-out (LOO) cross-validation on 101 brain neoplasms was achieved using the wrapper evaluator in combination with the Best First search algorithm and the KNN classifier and reached 96.9% when discriminating metastases from gliomas and 94.5% when discriminating high-grade from low-grade neoplasms.
Conclusions
A computer-assisted classification framework is developed and used for differential diagnosis of brain neoplasms based on MRI. The framework can achieve higher accuracy than most reported studies using MRI.
Similar content being viewed by others
References
Prayson RA, Agamanolis DP, Cohen ML, Estes ML (2000) Interobserver reproducibility among neuropathologists and surgical pathologists in fibrillary astrocytoma grading. J Neurol Sci 175(1): 33–39
Tate AR et al (2003) Automated classification of short echo time in in vivo 1H brain tumor spectra: a multicenter study. Magn Reson Med 49(1): 29–36
Majos C et al (2004) Brain tumor classification by proton MR spectroscopy: comparison of diagnostic accuracy at short and long TE. Am J Neuroradiol 25(10): 1696–1704
Huang Y, Lisboa PJG, El-Deredy W (2003) Tumour grading from magnetic resonance spectroscopy: a comparison of feature extraction with variable selection. Stat Med 22(1): 147–164
Lu C, Devos A, Suykens JAK, Arus C, Huffel S Van (2007) Bagging linear sparse Bayesian learning models for variable selection in cancer diagnosis. IEEE Trans Inf Technol Med 11(3): 338–346
Li G, Yang J, Ye C, Geng D (2006) Degree prediction of malignancy in brain glioma using support vector machines. Comput Biol Med 36(3): 313–325
Devos A et al (2005) The use of multivariate MR imaging intensities versus metabolic data from MR spectroscopic imaging for brain tumour classification. J Magn Reson 173(2): 218–228
Rajendran P, Madheswaran M (2009) An improved image mining technique for brain tumor classification using efficient classifier. Int J Comput Inf Secur 6(3)
Zacharaki EI, Wang S, Chawla S, Yoo DS, Wolf R, Melhem ER, Davatzikos C (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62: 1609–1618
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The WEKA data mining software: An update. SIGKDD Explor 11(1)
Liu H, Yu L (2005) Towards integrating feature selection algorithm for classification and clustering. IEEE Trans Knowl Data Eng 17(4): 491–502
Hall MA (2000) Correlation-based feature selection for discrete and numeric class machine learning. In: 17th international conference on machine learning (ICML):359–366
Dash M, Liu H (2003) Consistency-based search in feature selection. Artif Intell 151: 155–176
Kohavi R, John GH (1997) Wrappers for feature subset selection. Artif Intell 97: 273–324
Ghiselli EE (1964) Theory of psychological measurement. McGraw-Hill Book Co, New York
Xu L, Yan P, Chang T (1988) Best first strategy for feature selection. In: 9th international conference on pattern recognition, pp 706–708
Caruana R, Freitag D (1994) Greedy attribute selection. In: 11th international conference on machine learning, pp 28–36
Laguna M, Mart R (2003) Scatter search: methodology and implementations C. Kluwer, Dordrecht
Gandhi GM, Srivatsa SK (2010) Adaptive machine learning algorithm (AMLA) using J48 classifier for an NIDS environment. Adv Comput Sci Technol 3(3): 291–304
Cover TM, Hart PE (1967) Nearest neighbor pattern classification. Inst Electr Electron Eng Trans Inf Theory 13: 21–27
Demiroz G, Guvenir A (1997) Classification by voting feature intervals. In: 9th European conference on machine learning, pp 85–92
Chang C-C, Lin C-J (2001) LIBSVM: a library for support vector machines. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm
John GH, Langley P (1995) Estimating continuous distributions in bayesian classifiers. In: 11th conference on uncertainty in artificial intelligence, San Mateo, pp 338–345
Zacharaki EI, Wang S, Chawla S, Yoo DS, Wolf R, Melhem ER, Davatzikos C (2009) MRI-based classification of brain tumor type and grade using SVM-RFE. In: 6th IEEE International Symposis Biomedical Imaging (ISBI 2009), Boston, Massachusetts, USA
Huang Y-M, Du S-X (2005) weighted support vector machine for classification with uneven training class sizes. Int Conf Mach Learn Cybern 7: 4365–4369
Jolliffe IT (2002) Principal component analysis, series: springer series in statistics, 2nd edn. Springer, NY
Al-Okaili RN, Krejza J, Woo JH, Wolf RL, O’Rourke DM, Judy KD, Poptani H, Melhem ER (2007) Intraaxial brain masses: MR imaging–based diagnostic strategy—initial experience. Radiology 243: 539–550
Lev MH et al (2004) Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas. Am J Neuroradiol 25(2): 214–221
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zacharaki, E.I., Kanas, V.G. & Davatzikos, C. Investigating machine learning techniques for MRI-based classification of brain neoplasms. Int J CARS 6, 821–828 (2011). https://doi.org/10.1007/s11548-011-0559-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-011-0559-3