Abstract
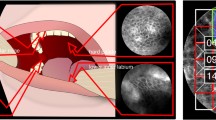
Squamous cell carcinoma (SCC) is the most common and malignant laryngeal cancer. An early-stage diagnosis is of crucial importance to lower patient mortality and preserve both the laryngeal anatomy and vocal-fold function. However, this may be challenging as the initial larynx modifications, mainly concerning the mucosa vascular tree and the epithelium texture and color, are small and can pass unnoticed to the human eye. The primary goal of this paper was to investigate a learning-based approach to early-stage SCC diagnosis, and compare the use of (i) texture-based global descriptors, such as local binary patterns, and (ii) deep-learning-based descriptors. These features, extracted from endoscopic narrow-band images of the larynx, were classified with support vector machines as to discriminate healthy, precancerous, and early-stage SCC tissues. When tested on a benchmark dataset, a median classification recall of 98% was obtained with the best feature combination, outperforming the state of the art (recall = 95%). Despite further investigation is needed (e.g., testing on a larger dataset), the achieved results support the use of the developed methodology in the actual clinical practice to provide accurate early-stage SCC diagnosis.

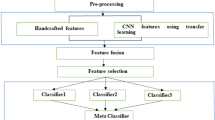
Workflow of the proposed solution. Patches of laryngeal tissue are pre-processed and feature extraction is performed. These features are used in the laryngeal tissue classification.







Similar content being viewed by others
References
McGuire S (2016) World cancer report 2014. Geneva, Switzerland: World Health Organization, international agency for research on cancer, WHO Press, 2015
Markou K, Christoforidou A, Karasmanis I, Tsiropoulos G, Triaridis S, Constantinidis I, Vital V, Nikolaou A (2013) Laryngeal cancer: Epidemiological data from Northern Greece and review of the literature. Hippokratia 17(4):313
Unger J, Lohscheller J, Reiter M, Eder K, Betz CS, Schuster M (2014) A noninvasive procedure for early-stage discrimination of malignant and precancerous vocal fold lesions based on laryngeal dynamics analysis. Cancer Res
Liang P, Cong Y, Guan M (2012) A computer-aided lesion diagnose method based on gastroscopeimage. In: IEEE international conference on information and automation. IEEE, pp 871– 875
Piazza C, Del Bon F, Peretti G, Nicolai P (2012) Narrow band imaging in endoscopic evaluation of the larynx. Current Opinion in Otolaryngology & Head and Neck Surgery 20(6):472–476
Isenberg JS, Crozier DL, Dailey SH (2008) Institutional and comprehensive review of laryngeal leukoplakia. Annals of Otology, Rhinology & Laryngology 117(1):74–79
Barbalata C, Mattos LS (2016) Laryngeal tumor detection and classification in endoscopic video. IEEE J Biomed Health Inform 20(1):322–332
Turkmen HI, Karsligil ME, Kocak I (2015) Classification of laryngeal disorders based on shape and vascular defects of vocal folds. Comput Biol Med 62:76–85
Moccia S, De Momi E, Guarnaschelli M, Savazzi M, Laborai A, Guastini L, Peretti G, Mattos L.S. (2017) Confident texture-based laryngeal tissue classification for early stage diagnosis support. J Med Imaging 4(3):034502
Nanni L, Ghidoni S, Brahnam S (2018) Appl Comput Inform. https://doi.org/10.1016/j.aci.2018.06.002
Zhang Y, Wirkert SJ, Iszatt J, Kenngott H, Wagner M, Mayer B, Stock C, Clancy NT, Elson DS, Maier-Hein L (2016) Tissue classification for laparoscopic image understanding based on multispectral texture analysis. In: Medical imaging image-guided procedures, robotic interventions, and modeling, vol 9786. International Society for Optics and Photonics, p 978619
Shen X, Sun K, Zhang S, Cheng S (2012) Lesion detection of electronic gastroscope images based on multiscale texture feature. In: IEEE International conference on signal processing, communication and computing. IEEE, pp 756–759
Misawa M, Kudo SE, Mori Y, Takeda K, Maeda Y, Kataoka S, Nakamura H, Kudo T, Wakamura K, Hayashi T, et al. (2017) Accuracy of computer-aided diagnosis based on narrow-band imaging endocytoscopy for diagnosing colorectal lesions: comparison with experts. International Journal of Computer Assisted Radiology and Surgery 12(5):757–766
Van Der Sommen F, Zinger S, Schoon EJ, et al. (2013) Computeraided detection of early cancer in the esophagus using HD endoscopy images. In: Medical imaging 2013: computer-aided diagnosis, vol 8670. International Society for Optics and Photonics, p 86700V
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) . IEEE Trans Med Imaging 35(5):1207. https://doi.org/10.1109/TMI.2016.2535865
Sirinukunwattana K, Raza SEA, Tsang Y, Snead DRJ, Cree IA, Rajpoot NM (2016) . IEEE Trans Med Imaging 35(5):1196. https://doi.org/10.1109/TMI.2016.2525803
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115 EP
Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, Peng L, Webster DR (2018) . Nature Biomed Eng 2(3):158. https://doi.org/10.1038/s41551-018-0195-0
Moccia S, Momi ED, Mattos LS (2017) Laryngeal dataset. https://doi.org/10.5281/zenodo.1003200
Jolliffe I (2011) Principal component analysis. https://doi.org/10.1007/978-3-642-04898-2_455
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: IEEE conference on computer vision and pattern recognition
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: IEEE conference on computer vision and pattern recognition
Szegedy C, Ioffe S, Vanhoucke V (2016) Computing Research Repository. arXiv:1602.07261
Burges CJ (1998) . Data Min Knowl Disc 2(2):121. https://doi.org/10.1023/A:1009715923555
Csurka G, Dance CR, Fan L, Willamowski J, Bray C (2004) Visual categorization with bags of keypoints. In: Workshop on statistical learning in computer vision, pp 1–22
Lin Y, Lv F, Zhu S, Yang M, Cour T, Yu K, Cao L, Huang T (2011) .. In: Conference on computer vision and pattern recognition. https://doi.org/10.1109/CVPR.2011.5995477, pp 1689–1696
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L (2015) . Int J Comput Vis 115(3):211. https://doi.org/10.1007/s11263-015-0816-y
Vivanti R, Joskowicz L, Lev-Cohain N, Ephrat A, Sosna J (2018) . Medical & Biological Engineering & Computing 56(9):1699. https://doi.org/10.1007/s11517-018-1803-6
Hatipoglu N, Bilgin G (2017) . Medical & Biological Engineering & Computing 55 (10):1829. https://doi.org/10.1007/s11517-017-1630-1
Moccia S, Foti S, Routray A, Prudente F, Perin A, Sekula RF, Mattos LS, Balzer JR, Fellows W, De Momi E, Riviere C (2018) Annals of Biomedical Engineering 46. https://doi.org/10.1007/s10439-018-2091-x
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Araújo, T., Santos, C.P., De Momi, E. et al. Learned and handcrafted features for early-stage laryngeal SCC diagnosis. Med Biol Eng Comput 57, 2683–2692 (2019). https://doi.org/10.1007/s11517-019-02051-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-019-02051-5