Abstract
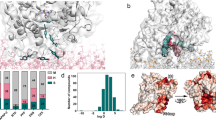
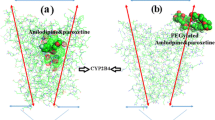
Drug-metabolizing enzymes, also known as cytochrome P450s, are a superfamily of hemoglobin responsible for metabolizing more than 90% clinical drugs. Cytochrome P450 2D6 (CYP2D6) is a significant member of cytochrome P450s for the reason of metabolizing about 20% clinical drugs. In this paper, molecular docking and molecular dynamic simulations are used to investigate the active site of CYP2D6, roles of essential amino acids within the active site and time-dependent protein energy changes. The results suggest that amino acids Glu216, Asp301, Ser304 and Ala305 in the active site are likely to form hydrogen bonding interactions with substrates; the benzene ring of Phe120 and aromatic ring in the substrates form Π-Π interactions. In addition, molecular dynamics simulations prove that the catalytic conformation of CYP2D6 without ligands can be obtained by their own atomic fluctuations. The impact of ligands on protein system energy and large conformational shift is not very large. Cytochrome P450s is known for their genetic polymorphisms, which will result in severe adverse drug reactions. Ideally, we hope to use molecular modeling to investigate the differences between the substrates of wild-type and mutants while they are bonded with drugs, and predict the drug metabolizing ability of mutants. Reduce the possibility for people taking drugs that they can not metabolize, therefore reduce the rate of adverse drug reactions, and eventually establish a platform of personalized drugs to largely benefit human health.
Similar content being viewed by others
References
Anzebacher P, Anzenbacherova E. Cytochrome P450 and metabolism of xenobiotics. Cell Mol Life Sci, 2001, 58: 737–747
Sono M, Roach M P, Coulter E D, et al. Heme-containing oxygenases. Chem Rev, 1996, 96: 2841–2887
Guengerich F P. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol, 2001, 14: 611–650
Williams P A, Cosme J. Mammalian microsomal cytochrome P450 monooxygenase: Structural adaptations for membrane binding and functional diversity. Mol Cell, 2000, 5: 121–131
Lewis D F, Jacobs M N, Dickins M. Compound lipophilicity for substrate binding to human P450s in drug metabolism. Drug Discov Today, 2004, 9: 530–537
Kirton S B, Murray C W, Verdonk M L, et al. Prediction of binding modes for ligands in the cytochrome P450 and other heme-containing proteins. Proteins, 2005, 58: 836–844
Stiborova M, Sopko B, Hodek P, et al. The binding of aristolochic acid I to the active site of human cytochromes P450 1A1 and 1A2 explains their potential to reductively activate this human carcinogen. Cancer Lett, 2005, 229: 193–204
Ito Y, Kondo H, Goldfarb P S, et al. Analysis of CYP2D6 substrate interactions by computational methods. J Mol Graph Model, 2008, 26: 947–956
Keizers P H, Lussenburg B M, de Graaf C, et al. Influence of phenylalanine 120 on cytochrome P450 2D6 catalytic selectivity regiospecificity: Crucial role in 7-methoxy-4-(aminomethyl)-coumarn metabolism. Biochem Pharmacol, 2004, 68: 2263–2271
Lussenburg B M, Keizers P H, de Graaf C, et al. The role of phenylalanine 483 in cytochrome P450 2D6 is strongly substrate dependent. Biochem Pharmacol, 2005, 70: 1253–1261
Rowland P, Blaney F E, Smyth M G, et al. Crystal structure of human cytochrome P450 2D6. J Biol Chem, 2006, 281: 7614–7622
Huey R, Morris G M, Olson A, et al. A semiempirical free energy force field with charge-based desolvation. J Comput Chem, 2007, 28: 1145–1152
Goodsell D S, Morris G M, Olson A J. Automated docking of flexible ligands: Applications of AutoDock. J Mol Recognition, 1996, 9: 15
Morris G M, Goodsell D S, Halliday R S, et al. Automated docking using a Lamarckian genetic algorithm and the empirical binding free energy function. J Comput Chem, 1998, 19: 1639–1662
Wang J F, Wei D Q, Chen C, et al. Molecular modeling of two CYP2C19 SNPs and its implications for peronalized drug design. Protein Pept Lett, 2008, 15: 27–32
Wang J F, Wei D Q, Zheng S Y, et al. 3D structure modeling of cytochrome P450 2C19 and its implication for personalized drug design. Biochem Biophys Res Commun, 2007, 355: 513–519
Wang J F, Wei D Q, Wang Y H, et al. Insight from modeling the 3D structure of NAD(P)H-dependent D-xylose reductase of Pichia stipitis and its binding interactions with NAD and NADP. Biochem Biophys Res Commun, 2007, 359: 323–329
Wang J F, Wei D Q, Du H L, et al. Molecular modeling studies on NADP-dependent of candida tropicalis strain xylose reductase. Open Bioinformatics J, 2008, 2: 89–96
Wei D Q, Du Q S, Sun H, et al. Insights from modeling the 3D structure of H5N1 influenza virus neuraminidase and its binding interactions with ligands. Biochem Biophys Res Commun, 2006, 344: 1048–1055
Berendsen H J C, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comp Phys Commun, 1995, 91: 43–56
Van Aalten D M F, Bywater R, Findlay J B, et al. PRODRG: A program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des, 1996, 10: 255–262
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Wang, J., Zhang, C., Wei, D. et al. Docking and molecular dynamics studies on CYP2D6. Chin. Sci. Bull. 55, 1877–1880 (2010). https://doi.org/10.1007/s11434-009-3697-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-009-3697-z