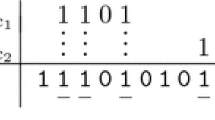
Abstract
Designing good or optimal seeds is a key factor for local homology search in bioinformatics. Continuous seeds have existed for nearly 20 years used by BLAST series programs. Recently, spaced seeds, which were introduced by PattenHunter program, were shown to be more sensitive and faster than continuous seeds under the same similarity level. However, there are 2 main disadvantages for space seeds: (i) It assumes that only matches and mismatches occur within seed alignments, but not insertions and deletions (indels); (ii) calculating optimal spaced seeds is an NP-hard problem. Introduction for indel seeds solved the first problem, but the second is getting much harder because of its higher exponential level. In this paper, we introduce an efficient way of designing good (even optimal) indel seeds under “indel overlap complexity” model, and it can be calculated in polynomial time. We calculate indel seeds from weight of 11 to 15. The result shows that indel seeds have higher sensitivities than spaced ones and our algorithm finds good indel seeds very quickly.
Similar content being viewed by others
References
Altschul S F, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol, 1990, 215: 403–410
Altschul S F, Madden T L, Schffer A A, et al. Gapped Blast and Psi-Blast: A new generation of protein database search programs. Nucleic Acids Res, 1997, 25: 3389–3402
Ma B, Tromp J, Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics, 2002, 18: 440–445
Li M, Ma B, Kisman D, et al. PatternHunter II: Highly sensitive and fast homology search. J Bioinform Comput Biol, 2004, 2: 164–175
Choi K P, Zeng F F, Zhang L. Good spaced seeds for homology search. Bioinformatics, 2004, 20: 1053–1059
Choi K P, Zhang L. Sensitivity analysis and efficient method for identifying optimal spaced seeds. J Comp System Sci, 2004, 68: 22–40
Keich U, Li M, Ma B, et al, On spaced seeds for similarity search. Discrete Appl Math, 2004, 138: 253–263
Zhang L. Superiority of spaced seeds for homology search. IEEE/ACM IEEE/ACM. Transact Comp Biol Bioinfor, 2007, 4: 496–505
Mark D, Gelfand Y, Benson G. Indel seeds for homology search. Bioinformatics, 2006, 22: 341–349
Li M, Ma B, Zhang L X. Superiority and Complexity of the Spaced Seeds. Proceedings of the 17th Symposium on Discrete Algorithms (SODA), 2006. 444–453
Ilie L, Ilie S. Long spaced seeds for finding similarities between biological sequences. Proceedings of BIOCOMP’07, 2007. 3–8
Ilie L, Ilie S. Multiple spaced seeds for homology search. Bioinformatics, 2007, 23: 2969–2977
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the National Natural Science Foundation of China (Grant No. 60671033) and the Doctor Research Foundation of the Ministry of Education China (Grant No. 20060614015)
About this article
Cite this article
Chen, K., Zhu, Q., Yang, F. et al. An efficient way of finding good indel seeds for local homology search. Chin. Sci. Bull. 54, 3837–3842 (2009). https://doi.org/10.1007/s11434-009-0531-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-009-0531-6