Abstract
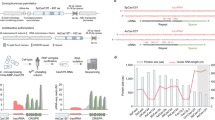
The length of the sgRNA-DNA complementary sequence is a key factor influencing the cleavage activity of Streptococcus pyogenes Cas9 (SpCas9) and its variants. The detailed mechanism remains unknown. Here, based on in vitro cleavage assays and base editing analysis, we demonstrate that reducing the length of this complementary region can confer nickase activity on SpCas9 and eSpCas9(1.1). We also show that these nicks are made on the target DNA strand. These properties encouraged us to develop a dual-functional system that simultaneously carries out double-strand DNA cleavage and C-to-T base conversions at separate targets. This system provides a novel tool for achieving trait stacking in plants.
Similar content being viewed by others
References
Casini, A., Olivieri, M., Petris, G., Montagna, C., Reginato, G., Maule, G., Lorenzin, F., Prandi, D., Romanel, A., Demichelis, F., et al. (2018). A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol 36, 265–271.
Chen, J.S., Dagdas, Y.S., Kleinstiver, B.P., Welch, M.M., Sousa, A.A., Harrington, L.B., Sternberg, S.H., Joung, J.K., Yildiz, A., and Doudna, J.A. (2017). Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410.
Cho, S.W., Kim, S., Kim, Y., Kweon, J., Kim, H.S., Bae, S., and Kim, J.S. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24, 132–141.
Doudna, J.A., and Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096.
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D.L., Wang, Z., Zhang, Z., Zheng, R., Yang, L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111, 4632–4637.
Fu, B.X.H., Smith, J.D., Fuchs, R.T., Mabuchi, M., Curcuru, J., Robb, G.B., and Fire, A.Z. (2019). Target-dependent nickase activities of the CRISPR-Cas nucleases Cpf1 and Cas9. Nat Microbiol 4, 888–897.
Fu, Y., Sander, J.D., Reyon, D., Cascio, V.M., and Joung, J.K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32, 279–284.
Gao, Y., and Zhao, Y. (2014). Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol 56, 343–349.
Hao, Y., Zong, W., Zeng, D., Han, J., Chen, S., Tang, J., Zhao, Z., Li, X., Ma, K., Xie, X., et al. (2020). Shortened snRNA promoters for efficient CRISPR/Cas-based multiplex genome editing in monocot plants. Sci China Life Sci https://doi.org/10.1007/s11427-019-1612-6.
Jiang, F., and Doudna, J.A. (2017). CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46, 505–529.
Kleinstiver, B.P., Pattanayak, V., Prew, M.S., Tsai, S.Q., Nguyen, N.T., Zheng, Z., and Joung, J.K. (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495.
Komor, A.C., Kim, Y.B., Packer, M.S., Zuris, J.A., and Liu, D.R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424.
Lee, J.K., Jeong, E., Lee, J., Jung, M., Shin, E., Kim, Y., Lee, K., Jung, I., Kim, D., Kim, S., et al. (2018). Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun 9, 3048.
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., Liu, J., Zhang, H., Liu, C., Ran, Y., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8, 14261.
Ma, H., Tu, L.C., Naseri, A., Huisman, M., Zhang, S., Grunwald, D., and Pederson, T. (2016). Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34, 528–530.
Puchta, H. (2018). Broadening the applicability of CRISPR/Cas9 in plants. Sci China Life Sci 61, 126–127.
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., Zhang, K., Liu, J., Xi, J.J., Qiu, J.L., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31, 686–688.
Slaymaker, I.M., Gao, L., Zetsche, B., Scott, D.A., Yan, W.X., and Zhang, F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88.
Szczelkun, M.D., Tikhomirova, M.S., Sinkunas, T., Gasiunas, G., Karvelis, T., Pschera, P., Siksnys, V., and Seidel, R. (2014). Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA 111, 9798–9803.
Tang, X., Liu, G., Zhou, J., Ren, Q., You, Q., Tian, L., Xin, X., Zhong, Z., Liu, B., Zheng, X., et al. (2018). A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol 19, 84.
Tsai, S.Q., Zheng, Z., Nguyen, N.T., Liebers, M., Topkar, V.V., Thapar, V., Wyvekens, N., Khayter, C., Iafrate, A.J., Le, L.P., et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197.
Wang, X., Lu, J., Lao, K., Wang, S., Mo, X., Xu, X., Chen, X., and Mo, B. (2019). Increasing the efficiency of CRISPR/Cas9-based gene editing by suppressing RNAi in plants. Sci China Life Sci 62, 982–984.
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., and Qiu, J.L. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32, 947–951.
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112, 3570–3575.
Zhang, D., Zhang, H., Li, T., Chen, K., Qiu, J.L., and Gao, C. (2017). Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol 18, 191.
Zhang, Q., Yin, K., Liu, G., Li, S., Li, M., and Qiu, J.L. (2020). Fusing T5 exonuclease with Cas9 and Cas12a increases the frequency and size of deletion at target sites. Sci China Life Sci https://doi.org/10.1007/s11427-020-1671-6.
Zhang, R., Liu, J., Chai, Z., Chen, S., Bai, Y., Zong, Y., Chen, K., Li, J., Jiang, L., and Gao, C. (2019). Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat Plants 5, 480–485.
Zhang, Y., Li, D., Zhang, D., Zhao, X., Cao, X., Dong, L., Liu, J., Chen, K., Zhang, H., Gao, C., et al. (2018). Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J 94, 857–866.
Zong, Y., Song, Q., Li, C., Jin, S., Zhang, D., Wang, Y., Qiu, J.L., and Gao, C. (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol 36, 950–953.
Zong, Y., Wang, Y., Li, C., Zhang, R., Chen, K., Ran, Y., Qiu, J.L., Wang, D., and Gao, C. (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35, 438–440.
Acknowledgements
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Precision Seed Design and Breeding, XDA24020102), the National Transgenic Science and Technology Program (2018ZX0801002B), the National Natural Science Foundation of China (31788103 and 31971370), the Chinese Academy of Sciences (QYZDY-SSW-SMC030), and the National Key R&D Program of China (2018YFA0900600, 2016YFD0100102-11, and 2016YFD0100605).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance and ethics The authors have filed a patent application based on the results reported in this paper. The patent does not restrict the research use of the methods in this article.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Fan, R., Chai, Z., Xing, S. et al. Shortening the sgRNA-DNA interface enables SpCas9 and eSpCas9(1.1) to nick the target DNA strand. Sci. China Life Sci. 63, 1619–1630 (2020). https://doi.org/10.1007/s11427-020-1722-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-020-1722-0