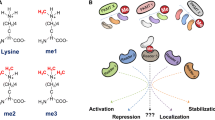
Abstract
Histone methylation is believed to play important roles in epigenetic memory in various biological processes. However, questions like whether the methylation marks themselves are faithfully transmitted into daughter cells and through what mechanisms are currently under active investigation. Previously, methylation was considered to be irreversible, but the recent discovery of histone lysine demethylases revealed a dynamic nature of histone methylation regulation on four of the main sites of methylation on histone H3 and H4 tails (H3K4, H3K9, H3K27 and H3K36). Even so, it is still unclear whether demethylases specific for the remaining two sites, H3K79 and H4K20, exist. Furthermore, besides histone proteins, the lysine methylation and demethylation also occur on non-histone proteins, which are probably subjected to similar regulation as histones. This review discusses recent progresses in protein lysine methylation regulation focusing on the above topics, while referring readers to a number of recent reviews for the biochemistry and biology of these enzymes
Similar content being viewed by others
References
Kouzarides T. Chromatin modifications and their function. Cell, 2007, 128(4): 693–705 17320507, 10.1016/j.cell.2007.02.005, 1:CAS:528:DC%2BD2sXis12ju7Y%3D
Lan F, Nottke A C, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol, 2008, 20(3): 316–325 18440794, 10.1016/j.ceb.2008.03.004, 1:CAS:528:DC%2BD1cXmvV2ls7k%3D
Taverna S D, Li H, Ruthenburg A J, et al. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol, 2007, 14(11): 1025–1040 17984965, 10.1038/nsmb1338, 1:CAS:528:DC%2BD2sXht1Kjs7vE
Bernstein B E, Mikkelsen T S, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell, 2006, 125(2): 315–326 16630819, 10.1016/j.cell.2006.02.041, 1:CAS:528:DC%2BD28Xkt1Oqur4%3D
Cui K, Zang C, Roh T Y, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell, 2009, 4(1): 80–93 19128795, 10.1016/j.stem.2008.11.011, 1:CAS:528:DC%2BD1MXhtVSks7s%3D
Verdel A, Jia S, Gerber S, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science, 2004, 303(5658): 672–676 14704433, 10.1126/science.1093686, 1:CAS:528:DC%2BD2cXmvVKmtQ%3D%3D
Yamada T, Fischle W, Sugiyama T, et al. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell, 2005, 20(2): 173–185 16246721, 10.1016/j.molcel.2005.10.002, 1:CAS:528:DC%2BD2MXht1SntrbJ
Rinn J L, Kertesz M, Wang J K, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 2007, 129(7): 1311–1323 17604720, 10.1016/j.cell.2007.05.022, 1:CAS:528:DC%2BD2sXotV2hsL4%3D
Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development, 2007, 134(2): 223–232 17185323, 10.1242/dev.02723, 1:CAS:528:DC%2BD2sXhvFGms7c%3D
Hansen K H, Bracken A P, Pasini D, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol, 2008, 10(11): 1291–1300 18931660, 10.1038/ncb1787, 1:CAS:528:DC%2BD1cXhtlais7jK
Schotta G, Lachner M, Sarma K, et al. A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev, 2004, 18(11): 1251–1262 15145825, 10.1101/gad.300704, 1:CAS:528:DC%2BD2cXkvVyrsb0%3D
Karachentsev D, Sarma K, Reinberg D, et al. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev, 2005, 19(4): 431–435 15681608, 10.1101/gad.1263005, 1:CAS:528:DC%2BD2MXhs1Ohs7o%3D
Kohlmaier A, Savarese F, Lachner M, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol, 2004, 2(7): E171 15252442, 10.1371/journal.pbio.0020171
Sims RJ 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol, 2008, 9(10): 815–820 18784729, 10.1038/nrm2502, 1:CAS:528:DC%2BD1cXhtFGhsLrN
Wang J, Hevi S, Kurash J K, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet, 2009, 41(1): 125–129 19098913, 10.1038/ng.268, 1:CAS:528:DC%2BD1cXhsFajtbbK
Chin H G, Estève P O, Pradhan M, et al. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res, 2007, 35(21): 7313–7323 17962312, 10.1093/nar/gkm726, 1:CAS:528:DC%2BD1cXhtFGltQ%3D%3D
Rathert P, Dhayalan A, Murakami M, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol, 2008, 4(6): 344–346 18438403, 10.1038/nchembio.88, 1:CAS:528:DC%2BD1cXlvFOisrs%3D
Goll M G, Bestor T H. Bestor, Eukaryotic cytosine methyltransferases. Annu Rev Biochem, 2005, 74: 481–514 15952895, 10.1146/annurev.biochem.74.010904.153721, 1:CAS:528:DC%2BD2MXpsVensLw%3D
Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol, 2006, 301: 179–201 16570848, 10.1007/3-540-31390-7_6, 1:CAS:528:DC%2BD28XivFeqtb8%3D
Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics, 2006, 1(2): 63–66 17965604, 10.4161/epi.1.2.2767
Hashimoto H, Horton J R, Zhang X, et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature, 2008, 455(7214): 826–829 18772888, 10.1038/nature07280, 1:CAS:528:DC%2BD1cXht1WktLnE
Bostick M, Kim J K, Estève P O, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science, 2007, 317(5845): 1760–1764 17673620, 10.1126/science.1147939, 1:CAS:528:DC%2BD2sXhtVGmsLbN
Fuks F, Hurd P J, Deplus R, et al. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res, 2003, 31(9): 2305–2312 12711675, 10.1093/nar/gkg332, 1:CAS:528:DC%2BD3sXjt1Ogt7o%3D
Ooi S K, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature, 2007, 448(7154): 714–717 17687327, 10.1038/nature05987, 1:CAS:528:DC%2BD2sXos1yksL8%3D
Smallwood A, Estève P O, Pradhan S, et al. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev, 2007, 21(10): 1169–1178 17470536, 10.1101/gad.1536807, 1:CAS:528:DC%2BD2sXls1aqu7w%3D
Schwartz Y B, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet, 2007, 8(1): 9–22 17173055, 10.1038/nrg1981, 1:CAS:528:DC%2BD28Xhtlakt7fM
Nishioka K, Rice J C, Sarma K, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell, 2002, 9(6): 1201–1213 12086618, 10.1016/S1097-2765(02)00548-8, 1:CAS:528:DC%2BD38Xlt1Sksbg%3D
Greeson N T, Sengupta R, Arida A R, et al. Di-methyl H4 lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J Biol Chem, 2008, 283(48): 33168–33174 18826944, 10.1074/jbc.M806857200, 1:CAS:528:DC%2BD1cXhsVSgt77I
Sanders S L, Portoso M, Mata J, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell, 2004, 119(5): 603–614 15550243, 10.1016/j.cell.2004.11.009, 1:CAS:528:DC%2BD2cXhtVGls7fM
Botuyan M V, Lee J, Ward I M, et al. Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell, 2006, 127(7): 1361–1373 17190600, 10.1016/j.cell.2006.10.043, 1:CAS:528:DC%2BD2sXmvVyksQ%3D%3D
Fraga M F, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet, 2005, 37(4): 391–400 15765097, 10.1038/ng1531, 1:CAS:528:DC%2BD2MXislCmurg%3D
Houston S I, McManus K J, Adams M M, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem, 2008, 283(28): 19478–19488 18480059, 10.1074/jbc.M710579200, 1:CAS:528:DC%2BD1cXotF2ksb8%3D
Trojer P, Li G, Sims R J 3rd, et al. L3MBTL1, a histone-methyla tion-dependent chromatin lock. Cell, 2007, 129(5): 915–928 17540172, 10.1016/j.cell.2007.03.048, 1:CAS:528:DC%2BD2sXmsl2qsLg%3D
Koga H, Matsui S, Hirota T, et al. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene, 1999, 18(26): 3799–3809 10445843, 10.1038/sj.onc.1202732, 1:CAS:528:DyaK1MXks1Omtr8%3D
Kotake Y, Cao R, Viatour P, et al. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev, 2007, 21(1): 49–54 17210787, 10.1101/gad.1499407, 1:CAS:528:DC%2BD2sXmsFyltw%3D%3D
Kuzmichev A, Jenuwein T, Tempst P, et al. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell, 2004, 14(2): 183–193 15099518, 10.1016/S1097-2765(04)00185-6, 1:CAS:528:DC%2BD2cXjvVyisb8%3D
Kuzmichev A, Margueron R, Vaquero A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA, 2005, 102(6): 1859–1864 15684044, 10.1073/pnas.0409875102, 1:CAS:528:DC%2BD2MXhvFGrtLo%3D
Trojer P, Zhang J, Yonezawa M, et al. Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9A/KMT1C and the jumonji domain containing JMJD2/KDM4 proteins. J Biol Chem, 2009, doi: 10.1074/jbc.M807818200
Lachner M, O’Carroll D, Rea S, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 2001, 410(6824): 116–120 11242053, 10.1038/35065132, 1:CAS:528:DC%2BD3MXhvVSltr8%3D
Aagaard L, Laible G, Selenko P, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J, 1999, 18(7): 1923–1938 10202156, 10.1093/emboj/18.7.1923, 1:CAS:528:DyaK1MXivVyks7o%3D
Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol, 2004, 5(4): 296–304 15071554, 10.1038/nrm1355, 1:CAS:528:DC%2BD2cXivV2itr8%3D
Collins R E, Northrop J P, Horton J R, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol, 2008, 15(3): 245–250 18264113, 10.1038/nsmb.1384, 1:CAS:528:DC%2BD1cXislKmtb8%3D
Shi Y, Whetstine J R. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell, 2007, 25(1): 1–14 17218267, 10.1016/j.molcel.2006.12.010, 1:CAS:528:DC%2BD2sXhtFSntbg%3D
Klose R J, Kallin E M, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet, 2006, 7(9): 715–727 16983801, 10.1038/nrg1945, 1:CAS:528:DC%2BD28Xot1Ggu74%3D
Lan F, Bayliss P E, Rinn J L, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature, 2007, 449(7163): 689–694 17851529, 10.1038/nature06192, 1:CAS:528:DC%2BD2sXhtFChurfJ
Lan F, Collins R E, De Cegli R, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature, 2007, 448(7154): 718–722 17687328, 10.1038/nature06034, 1:CAS:528:DC%2BD2sXos1yksLk%3D
Huang Y, Greene E, Murray-Stewart T, et al. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci USA, 2007, 104(19): 8023–8028 17463086, 10.1073/pnas.0700720104, 1:CAS:528:DC%2BD2sXmtFCiurg%3D
Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell, 2007, 129(4): 823–837 17512414, 10.1016/j.cell.2007.05.009, 1:CAS:528:DC%2BD2sXmtFKjsro%3D
Iwase S, Lan F, Bayliss P, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell, 2007, 128(6): 1077–1088 17320160, 10.1016/j.cell.2007.02.017, 1:CAS:528:DC%2BD2sXkslGms7w%3D
Whetstine J R, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell, 2006, 125(3): 467–481 16603238, 10.1016/j.cell.2006.03.028, 1:CAS:528:DC%2BD28XkslCjtLo%3D
Trojer P, Reinberg D. Beyond histone methyl-lysine binding: how malignant brain tumor (MBT) protein L3MBTL1 impacts chromatin structure. Cell Cycle, 2008, 7(5): 578–585 18256536, 1:CAS:528:DC%2BD1cXlvFensL0%3D
Pesavento J J, Yang H, Kelleher N L, et al. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol, 2008, 28(1): 468–486 17967882, 10.1128/MCB.01517-07, 1:CAS:528:DC%2BD1cXktVaruw%3D%3D
Kachirskaia I, Shi X, Yamaguchi H, et al. Role for 53BP1 Tudor domain recognition of p53 dimethylated at lysine 382 in DNA damage signaling. J Biol Chem, 2008, 283(50): 34660–34666 18840612, 10.1074/jbc.M806020200, 1:CAS:528:DC%2BD1cXhsVGqtrnJ
Shi X, Kachirskaia I, Yamaguchi H, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell, 2007, 27(4): 636–646 17707234, 10.1016/j.molcel.2007.07.012, 1:CAS:528:DC%2BD2sXhtVSmtL3E
Luger K, Mäder A W, Richmond R K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature, 1997, 389(6648): 251–260 9305837, 10.1038/38444, 1:CAS:528:DyaK2sXmtFGisrc%3D
Feng Q, Wang H, Ng H H, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol, 2002, 12(12): 1052–1058 12123582, 10.1016/S0960-9822(02)00901-6, 1:CAS:528:DC%2BD38XlsVyjtLY%3D
Janzen C J, Hake S B, Lowell J E, et al. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell, 2006, 23(4): 497–507 16916638, 10.1016/j.molcel.2006.06.027, 1:CAS:528:DC%2BD28Xpt1Sntbg%3D
San-Segundo P A, Roeder G S. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell, 2000, 11(10): 3601–3615 11029058, 1:CAS:528:DC%2BD3cXnslWntLY%3D
Ooga M, Inoue A, Kageyama S, et al. Changes in H3K79 methylation during preimplantation development in mice. Biol Reprod, 2008, 78(3): 413–424 18003948, 10.1095/biolreprod.107.063453, 1:CAS:528:DC%2BD1cXisFSltbo%3D
Fang J, Feng Q, Ketel C S, et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol, 2002, 12(13): 1086–1099 12121615, 10.1016/S0960-9822(02)00924-7, 1:CAS:528:DC%2BD38XlsVyjsLg%3D
McGinty R K, Kim J, Chatterjee C, et al. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature, 2008, 453(7196): 812–816 18449190, 10.1038/nature06906, 1:CAS:528:DC%2BD1cXmvVGmsLw%3D
Kuzmichev A, Nishioka K, Erdjument-Bromage H, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev, 2002, 16(22): 2893–2905 12435631, 10.1101/gad.1035902, 1:CAS:528:DC%2BD38XovVejsLY%3D
Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science, 2002, 298(5595): 1039–1043 12351676, 10.1126/science.1076997, 1:CAS:528:DC%2BD38Xot12rs7g%3D
Czermin B, Melfi R, McCabe D, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell, 2002, 111(2): 185–196 12408863, 10.1016/S0092-8674(02)00975-3, 1:CAS:528:DC%2BD38Xotlahsr8%3D
Müller J, Hart C M, Francis N J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell, 2002, 111(2): 197–208 12408864, 10.1016/S0092-8674(02)00976-5
Lin C H, Li B, Swanson S, et al. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell, 2008, 32(5): 696–706 19061644, 10.1016/j.molcel.2008.11.008, 1:CAS:528:DC%2BD1cXhsFSntrrJ
Lee M G, Wynder C, Cooch N, et al. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature, 2005, 437(7057): 432–435 16079794, 1:CAS:528:DC%2BD2MXpvFOrtL8%3D
Shi Y J, Matson C, Lan F, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell, 2005, 19(6): 857–864 16140033, 10.1016/j.molcel.2005.08.027, 1:CAS:528:DC%2BD2MXhtVyhtbzK
Huang J, Perez-Burgos L, Placek B J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature, 2006, 444(7119): 629–632 17108971, 10.1038/nature05287, 1:CAS:528:DC%2BD28Xht1KjurjP
Huang J, Sengupta R, Espejo A B, et al. p53 is regulated by the lysine demethylase LSD1. Nature, 2007, 449(7158): 105–108 17805299, 10.1038/nature06092, 1:CAS:528:DC%2BD2sXpvFWrs7w%3D
Ivanov G S, Ivanova T, Kurash J, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol, 2007, 27(19): 6756–6769 17646389, 10.1128/MCB.00460-07, 1:CAS:528:DC%2BD2sXhtFSktL%2FL
Chuikov S, Kurash J K, Wilson J R, et al. Regulation of p53 activity through lysine methylation. Nature, 2004, 432(7015): 353–360 15525938, 10.1038/nature03117, 1:CAS:528:DC%2BD2cXpvVCjtbo%3D
Couture J F, Collazo E, Hauk G, et al. Structural basis for the methylation site specificity of SET7/9. Nat Struct Mol Biol, 2006, 13(2): 140–146 16415881, 10.1038/nsmb1045, 1:CAS:528:DC%2BD28XhtFajsb4%3D
Kouskouti A, Scheer E, Staub A, et al. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell, 2004, 14(2): 175–182 15099517, 10.1016/S1097-2765(04)00182-0, 1:CAS:528:DC%2BD2cXjvVyisb4%3D
Subramanian K, Jia D, Kapoor-Vazirani P, et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell, 2008, 30(3): 336–347 18471979, 10.1016/j.molcel.2008.03.022, 1:CAS:528:DC%2BD1cXmtFaktLs%3D
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lan, F., Shi, Y. Epigenetic regulation: methylation of histone and non-histone proteins. SCI CHINA SER C 52, 311–322 (2009). https://doi.org/10.1007/s11427-009-0054-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-009-0054-z