Abstract
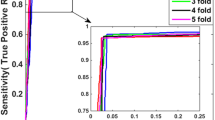
This paper develops sequence-based methods for identifying novel protein-protein interactions (PPIs) by means of support vector machines (SVMs). The authors encode proteins ont only in the gene level but also in the amino acid level, and design a procedure to select negative training set for dealing with the training dataset imbalance problem, i.e., the number of interacting protein pairs is scarce relative to large scale non-interacting protein pairs. The proposed methods are validated on PPIs data of Plasmodium falciparum and Escherichia coli, and yields the predictive accuracy of 93.8% and 95.3%, respectively. The functional annotation analysis and database search indicate that our novel predictions are worthy of future experimental validation. The new methods will be useful supplementary tools for the future proteomics studies.
Similar content being viewed by others
References
J. Wang, S. Zhang, Y. Wang, et al., Disease-aging network reveals significant roles of aging genes in connecting genetic diseases, PLoS Computational Biology, 2009, 5(9): e1000521.
S. Fields and O. Song, A novel genetic system to detect protein-protein interactions, Nature, 1989, 340: 245–246.
T. Ito, T. Chiba, R. Ozawa, et al., A comprehensive two-hybrid analysis to explore the yeast protein interactome, Proceedings of the National Academy of Sciences, 2001, 98: 4569–4574.
A. C. Gavin, M. Boche, R. Krause, et al., Functional organization of the yeast proteome by systematic analysis of protein complexes, Nature, 2002, 415: 141–147.
Y. Ho, A. Gruhler, A. Heilbut, et al., Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry, Nature, 2002, 415: 180–183.
H. Zhu, M. Bilgin, R. Bangham, et al., Global analysis of protein activities using proteome chips, Science, 2001, 193: 2101–2105.
Y. Z. Guo, L. Z. Yu, Z. N. Wen, and M. L. Li, Using support vector machine combined with auto covariance to predict protein-protein interactions from protein sequences, Nucleic Acids Research, 2008, 36: 3025–3030.
S. Martin, D. Roe, and J. L. Faulon, Predicting protein-protein interactions using signature products, Bioinformatics, 2005, 21: 218–226.
K. C. Chou and Y. D. Cai, Predicting protein-protein interactions from sequences in a hybridization space, Journal of Proteome Research, 2006, 5: 316–322.
R. Jansen, H. J. Bussemaker, and M. Gerstein, Revisiting the codon adaptation index from a wholegenome perspective: Analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models, Nucleic Acids Research, 2003, 31: 2242–2251.
K. A. Dittmar, M. A. Sorensen, J. Elf, et al., Selective charging of tRNA isoacceptors induced by amino-acid starvation, EMBO Reports, 2005, 6: 151–157.
H. S. Najafabadi and R. Salavati, Sequence-based prediction of protein-protein interactions by means of codon usage, Genome Biology, 2008, 9: R87–R95.
J. W. Shen, J. Zhang, X. M. Luo, et al., Predicting protein-protein interactions based only on sequences information, Proceedings of the National Academy of Sciences, 2007, 104: 4337–4341.
B. Schökopf and A. J. Smola, Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond, MIT Press, Cambridge, MA, 2002.
B. Schökopf, K. Tsuda, and J. P. Vert, Kernel Methods in Computational Biology, MIT Press, Cambridge, MA, 2004, 71–92.
S. Kerrien, Y. Alam-Faruque, B. Aranda, et al., IntAct-open source resource for molecular interaction data, Nucleic Acids Research, 2007, 35: D561–D565.
L. Salwinski, C. S. Miller, A. J. Smith, et al., The database of interacting proteins: 2004 update, Nucleic Acids Research, 2004, 32: D449–D451.
G. D. Bader, I. Donaldson, C. Wolting, et al., BIND: The Biomolecular Interaction Network Database, Nucleic Acids Research, 2003, 31: 248–250.
G. R. Mishra, M. Suresh, K. Kumaran, et al., Human protein reference database-2006 update, Nucleic Acids Research, 2006, 34: 411–414.
Y. C. Wang, J. G. Wang, Z. X. Yang, et al., Prediction of protein-protein interaction based only on coding sequences, Proceedings of the 8th International Symposium on Optimization and Systems Biology, Zhangjiajie, 2009, 151–158.
C. W. Hsu, C. C. Chang, and C. J. Lin, A practical guide to Support Vector Classfication, 2007, URL: http://www.csie.ntu.edu.tw/cjlin.
M. Gribskov and N. L. Robinson, Use of receiver operating characteristic (roc) analysis to evaluate sequence matching, Computers and Chemistry, 1996, 20: 25–33.
J. Platt, Probabilistic outputs for support vector machines and comparison to regularized likelihood methods, Advances in Large Margin Classifiers, 1999: 61–74.
D. J. LaCount, M. Vignali, R. Chettier, et al., A protein interaction network of the malaria parasite Plasmodium falciparum, Nature, 2005, 10: 103–107.
G. D. Bader and C. W. Hogue, Analyzing yeast protein-protein interaction data obtained from different sources, Nature Biotechnology, 2002, 20: 991–997.
A. Kumar and M. Snyder, Protein complexes take the bait, Nature, 2002, 415: 123–124.
C. Hertz-Fowler, C. S. Peacock, V. Wood, et al., GeneDB: A resource for prokaryotic and eukaryotic organisms, Nucleic Acids Research, 2004, 32: D339–D343.
C. Aurrecoechea, J. Brestelli, B. P. Brunk, et al., PlasmoDB: A functional genomic database for malaria parasites, Nucleic Acids Research, 2009, 37: D539–D543.
C. Su, J. M. Peregrin-Alvarez, G. Butland, et al., Bacteriome.org-an integrated protein interaction database for E. coli, Nucleic Acids Research, 2008, 36: D632–D636.
I. M. Keseler, C. Bonavides-Martínez, J. Collado-Vides, et al., EcoCyc: A comprehensive view of Escherichia coli biology, Nucleic Acids Research, 2009, 37: D464–D470.
E. Andres Leon, I. Ezkurdia, B. García, et al., EcID. A database for the inference of functional interactions in E. coli, Nucleic Acids Research, 2009, 37: D629–D635.
A. Ben-Hur and W. S. Noble, Kernel methods for predicting protein-protein interactions, Bioinformatics, 2005, 21: i38–i46.
K. C. Chou, Prediction of protein cellular attributes using pseudo-amino acid composition, Proteins: Structure, Function, and Genetics, 2001, 43: 246–255.
K. C. Chou, Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes, Bioinformatics, 2005, 21: 10–19.
G. R. Lanckriet, M. Deng, N. Cristianini, et al., Kernel-based data fusion and its application to protein function prediction in yeast, Pacific Symposium on Biocomputing, 2004.
Y. Guan, C. Myers, D. Hess, et al., Predicting gene function in a hierarchical context with an ensemble of classifiers, Genome Biology, 2008, 9(S3).
B. Li, J. Hu, K. Hirasawa, et al., Support vector machine with fuzzy decision-making for realworld data classification, IEEE World Congress on Computational Intelligence, Int. Joint Conf. on Neural Networks, Canada, 2006.
R. Jayadeva Khemchandani and S. Chandra, Twin support vector machines for pattern classification, IEEE Transactions on Pattern Analysis and Machine Intelligence, 2007, 29: 905–910.
S. Ghorai, A. Mukherjee, and P. K. Dutta, Nonparallel plane proximal classifier, Signal Processing, 2008, 89: 510–522.
Author information
Authors and Affiliations
Corresponding author
Additional information
This research is supported by the Key Project of the National Natural Science Foundation of China under Grant No. 10631070, the National Natural Science Foundation of China under Grant Nos. 10801112, 10971223, 11071252, and the Ph.D Graduate Start Research Foundation of Xinjiang University Funded Project under Grant No. BS080101.
Rights and permissions
About this article
Cite this article
Wang, Y., Wang, J., Yang, Z. et al. Sequence-based protein-protein interaction prediction via support vector machine. J Syst Sci Complex 23, 1012–1023 (2010). https://doi.org/10.1007/s11424-010-0214-z
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11424-010-0214-z