Abstract
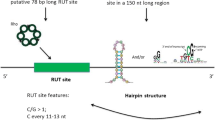
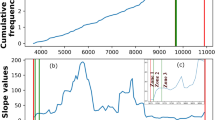
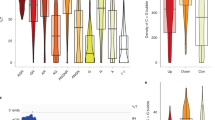
A new method for intrinsic terminator prediction based on Rnall, an RNA local secondary structure prediction algorithm developed recently, and two U-tail score schemas are developed. By optimizing three parameters (thermodynamic energy of RNA hairpin structure, U-tail T weight, and U-tail hybridization energy), the method can recognize 92.25% of known terminators while rejecting 98.48% of predicted RNA local secondary structures in coding regions (negative control) as false intrinsic terminators in E. coli. This method was applied to scan the genome of Synechococcus sp. WH8102, and we predicted 266 intrinsic terminators, which included 232 protein-coding genes, 12 tRNA genes, and 3 rRNA genes. About 17% of these terminators are located at the end of operons. It is also identified 8 pairs of bio-directional terminators. The method for intrinsic terminator prediction has been incorporated into Rnall, which is available at http://digbio.missouri.edu/∼wanx/Rnall/.
Similar content being viewed by others
References
Richardson J P. Loading rho to terminate transcription. Cell, 2003, 114: 157–159.
Henkin T M. Control of transcription termination in prokaryotes. Annu. Rev. Genet., 1996, 30: 35–57.
d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol., 1990, 216: 835–858.
Yarnell W S, Roberts J W. Mechanism of intrinsic transcription termination and antitermination. Science, 1999, 284: 611–615.
Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell, 1999, 3: 495–504.
Grundy F J et al. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: Disparity between conservation and functional requirements. Nucleic Acids Res., 2002, 30: 1646–1655.
Brendel V, Trifonov E N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res., 1984, 12: 4411–4427.
Ermolaeva M D et al. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol., 2000, 301: 27–33.
Lesnik E A et al. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res., 2001, 29: 3583–3594.
Unniraman S, Prakash R, Nagaraja V. Conserved economics of transcription termination in eubacteria. Nucleic Acids Res., 2002, 30: 675–684.
Palenik B et al. The genome of a motile marine Synechococcus. Nature, 2003, 424: 1037–1042.
Hess W R. Genome analysis of marine photosynthetic microbes and their global role. Curr. Opin. Biotechnol., 2004, 15: 191–198.
Scanlan D J. Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microb. Physiol., 2003, 47: 1–64.
Chen X, Su Z, Dam P, Palenik B, Xu Y, Jiang T. Operon prediction by comparative genomics: An application to the Synechococcus sp. WH8102 genome. Nucleic Acids Res., 2004, 32: 2147–2157.
Wan X-F, Xu D. Rnall: A novel algorithm for RNA local secondary structure prediction. (submitted)
Altschul S F et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res., 1997, 25: 3389–3402.
Mathews D H et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. In Proc. Natl. Acad. Sci. U.S.A., 2004, 101: 7287–7292.
Mathews D H et al. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 1999, 288: 911–940.
Sugimoto N et al. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry, 1995, 34: 11211–11216.
Sun Z, Xia X, Guo Q, Xu D. Protein structure prediction in a 210-type lattice model: Parameter optimization in the genetic algorithm using orthogonal array. J. Protein Chem., 1999, 18: 39–46.
Wang L, Trawick J D, Yamamoto R, Zamudio C. Genome-wide operon prediction in Staphylococcus aureus. Nucleic Acids Res., 2004, 32: 3689–3702.
Washio T, Sasayama J, Tomita M. Analysis of complete genomes suggests that many prokaryotes do not rely on hairpin formation in transcription termination. Nucleic Acids Res., 1998, 26: 5456–5463.
Yanofsky C. Transcription attenuation: Once viewed as a novel regulatory strategy. J Bacteriol., 2000, 182: 1–8.
Ingham C J, Hunter I S, Smith M C. Rho-independent terminators without 3′ poly-U tails from the early region of actinophage oC31. Nucleic Acids Res., 1995, 23: 370–376.
Reynolds R, Chamberlin M J. Parameters affecting transcription termination by Escherichia coli RNA. II. Construction and analysis of hybrid terminators. J Mol Biol., 1992, 224: 53–63.
Abe H, Aiba H. Differential contributions of two elements of rho-independent terminator to transcription termination and mRNA stabilization. Biochimie, 1996, 78: 1035–1042.
Unniraman S, Prakash R, Nagaraja V. Alternate paradigm for intrinsic transcription termination in eubacteria. J Biol Chem., 2001, 276: 41850–41855.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work has been supported by the US Department of Energy's Genomes to Life program (http://www.doegenomestolife.org) under project, “Carbon Sequestration in Synechococcus Sp.: From Molecular Machines to Hierarchical Modeling“ (http://www.genomes-to-life.org), and by National Science Foundation (EIA-0325386). The research used supercomputer resources of the Center for Computational Sciences at Oak Ridge National Laboratory, which is supported by the Office of Science of the Department of Energy under Contract DE-AC05-00OR22725
Xiu-Feng Wan is a postdoctoral researcher in the Digital Biology Laboratory, Department of Computer Science, University of Missouri, Columbia, MO. In August of 2005, he will be an assistant professor of bioinformatics and computational biology at the Department of Microbiology, Miami University, Oxford, OH. His lab will perform both computational studies and wet bench validation. His research interests include computational modeling of infectious diseases (Avian Influenza, SARS, and HIV/AIDS), transcriptional regulatory motif (both DNA and RNA motif) identification, regulatory network construction using microarray data, and immunological pathway modeling using functional genomics. He received his B.S. degree in veterinary medicine from Jiangxi Agricultural University (1995), M.S. degree in avian medicine from South China Agricultural University (1998), and Ph.D. degree in veterinary medicine and minor in biochemistry and molecular biology from Mississippi State University (2002). He also received his M.Sc. in computer science from Mississippi State University (2002). He obtained his postdoc trainings from Oak Ridge National Laboratory (2002--2003) and University of Missouri, Columbia (2004--2005).
Dr. Dong Xu is a James C. Dowell Associate Professor and director of Digital Biology Laboratory in the Computer Science Department, University of Missouri, Columbia. He obtained his Ph.D. from the University of Illinois, Urbana-Champaign in 1995 and did two-year postdoctoral work at National Cancer Institute. He was a staff scientist at Oak Ridge National Laboratory until August 2003 before joining University of Missouri. Over the past fourteen years, he has done active research in many areas of computational biology and bioinformatics with more than 90 scientific papers. He is a recipient of the year 2001 R&D 100 award, a prestigious international award sponsored by Research & Development magazine that honors the 100 most significant new technical products of the previous year, for developing “Protein Structure Prediction and Evaluation Computer Toolkit (PROSPECT)”. He also received 2003 Award of Excellence in Technology Transfer from The Federal Laboratory Consortium for developing gene expression analysis package EXCAVATOR. He has received research grants from the Department of Energy, the National Science Foundation, the U.S. Department of Agriculture, National Institute of Health, Ceres Inc., and Monsanto Research Funds. He has served on the Program Committee or as a Session Chair for major bioinformatics conferences, including IEEE Computer Society Bioinformatics Conference (CSB), Annual Conference of Intelligent Systems for Molecular Biology (ISMB), Annual International Conference IEEE Engineering in Medicine and Biology Society (EMBS), and European Conference on Computational Biology (ECCB). He has been a member of the Editorial Board of “Current Protein and Peptide Science” since January 2000.
Rights and permissions
About this article
Cite this article
Wan, XF., Xu, D. Intrinsic Terminator Prediction and Its Application in Synechococcus sp. WH8102. J Comput Sci Technol 20, 465–482 (2005). https://doi.org/10.1007/s11390-005-0465-7
Revised:
Issue Date:
DOI: https://doi.org/10.1007/s11390-005-0465-7