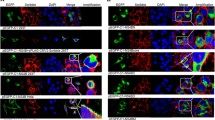
Abstract
The ARFP/F protein is synthesized from the +1 reading frame of the hepatitis C virus (HCV) core protein gene. The function of this protein remains unknown. To study the function of the HCV ARFP/F protein, we have conducted the yeast two-hybrid screening experiment to identify cellular proteins that may interact with the ARFP/F protein. MM-1, a c-Myc interacting protein, was found to interact with HCV ARFP/F protein in this experiment. The physical interaction between ARFP/F and MM-1 proteins was further confirmed by the GST pull-down assay, the co-immunoprecipitation assay and confocal microscopy. As MM-1 can inhibit the gene transactivation activity of c-Myc, we have conducted further analysis to examine the possible effect of the ARFP/F protein on c-Myc. Our results indicate that the HCV ARFP/F protein can enhance the gene trans-activation activity of c-Myc, apparently by antagonizing the inhibitory effect of MM-1. The ability of the ARFP/F protein to enhance the activity of c-Myc raises the possibility that ARFP/F protein might play a role in hepatocellular transformation in HCV patients.







Similar content being viewed by others
References
Reed KE, Rice CM (2000) Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol 242:55–84
Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM (1993) Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol 67:1385–1395
Lin C, Lindenbach BD, Pragai BM, McCourt DW, Rice CM (1994) Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol 68:5063–5073
Rosenberg S (2001) Recent advances in the molecular biology of hepatitis C virus. J Mol Biol 313:451–464
Santolini E, Migliaccio G, La Monica N (1994) Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol 68:3631–3641
Hussy P, Langen H, Mous J, Jacobsen H (1996) Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology 224:93–104
Ray RB, Lagging LM, Meyer K, Steele R, Ray R (1995) Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res 37:209–220
Chen CM, You LR, Hwang LH, Lee YH (1997) Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-beta receptor modulates the signal pathway of the lymphotoxin-beta receptor. J Virol 71:9417–9426
Heim MH, Moradpour D, Blum HE (1999) Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol 73:8469–8475
Yoshida T, Hanada T, Tokuhisa T, Kosai K, Sata M, Kohara M, Yoshimura A (2002) Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J Exp Med 196:641–653
Tsutsumi T, Suzuki T, Shimoike T, Suzuki R, Moriya K, Shintani Y, Fujie H, Matsuura Y, Koike K, Miyamura T (2002) Interaction of hepatitis C virus core protein with retinoid X receptor alpha modulates its transcriptional activity. Hepatology 35:937–946
Honda M, Kaneko S, Shimazaki T, Matsushita E, Kobayashi K, Ping LH, Zhang HC, Lemon SM (2000) Hepatitis C virus core protein induces apoptosis and impairs cell-cycle regulation in stably transformed Chinese hamster ovary cells. Hepatology 31:1351–1359
Ray RB, Meyer K, Steele R, Shrivastava A, Aggarwal BB, Ray R (1995) Inhibition of tumor necrosis factor (TNF-alpha)-mediated apoptosis by hepatitis C virus core protein. J Biol Chem 273:2256–2259
Ruggieri A, Harada T, Matsuura Y, Miyamura T (1997) Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology 229:68–76
Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, Miyamura T, Koike K (1997) Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol 78(Pt 7):1527–1531
Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K (1998) The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med 4:1065–1075
Walewski JL, Keller TR, Stump DD, Branch AD (2001) Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. Rna 7:710–721
Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J (2001) Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. Embo J 20:3840–3848
Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P (2002) Alternate translation occurs within the core coding region of the hepatitis C viral genome. J Biol Chem 277:17713–17721
Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL (2005) The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis 25:105–117
Komurian-Pradel F, Rajoharison A, Berland JL, Khouri V, Perret M, Van Roosmalen M, Pol S, Negro F, Paranhos-Baccala G (2004) Antigenic relevance of F protein in chronic hepatitis C virus infection. Hepatology 40:900–909
Bain C, Parroche P, Lavergne JP, Duverger B, Vieux C, Dubois V, Komurian-Pradel F, Trepo C, Gebuhrer L, Paranhos-Baccala G, Penin F, Inchauspe G (2004) Memory T-cell-mediated immune responses specific to an alternative core protein in hepatitis C virus infection. J Virol 78:10460–10469
Vassilaki N, Mavromara P (2003) Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core + 1 coding open reading frame. J Biol Chem 278:40503–40513
Boulant S, Becchi M, Penin F, Lavergne JP (2003) Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J Biol Chem 278:45785–45792
Baril M, Brakier-Gingras L (2005) Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucl Acids Res 33:1474–1486
Xu Z, Choi J, Lu W, Ou JH (2003) Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum. J Virol 77:1578–1583
Roussel J, Pillez A, Montpellier C, Duverlie G, Cahour A, Dubuisson J, Wychowski C (2003) Characterization of the expression of the hepatitis C virus F protein. J Gen Virol 84:1751–1759
Basu A, Steele R, Ray R, Ray RB (2004) Functional properties of a 16 kDa protein translated from an alternative open reading frame of the core-encoding genomic region of hepatitis C virus. J Gen Virol 85:2299–2306
Mori K, Maeda Y, Kitaura H, Taira T, Iguchi-Ariga SM, Ariga H (1998) MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc. J Biol Chem 273:29794–29800
Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM (1997) Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570–574
Fuerst TR, Niles EG, Studier FW, Moss B (1986) Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA 83:8122–8126
Ma HC, Ke CH, Hsieh TY, Lo SY (2002) The first hydrophobic domain of the hepatitis C virus E1 protein is important for interaction with the capsid protein. J Gen Virol 83:3085–3092
Lo SY, Selby M, Tong M, Ou JH (1994) Comparative studies of the core gene products of two different hepatitis C virus isolates: two alternative forms determined by a single amino acid substitution. Virology 199:124–131
Satou A, Hagio Y, Taira T, Iguchi-Ariga SM, Ariga H (2004) Repression of the c-fms gene in fibroblast cells by c-Myc-MM-1-TIF1beta complex. FEBS Lett 572:211–215
Sakamuro D, Prendergast GC (1999) New Myc-interacting proteins: a second Myc network emerges. Oncogene 18:2942–2954
Wang Y, Wu MC, Sham JS, Zhang W, Wu WQ, Guan XY (2002) Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer 95:2346–2352
Peng SY, Lai PL, Hsu HC (1993) Amplification of the c-myc gene in human hepatocellular carcinoma: biologic significance. J Formos Med Assoc 92:866–870
Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW (2004) MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431:1112–1117
Transy C, Fourel G, Robinson WS, Tiollais P, Marion PL, Buendia MA (1992) Frequent amplification of c-myc in ground squirrel liver tumors associated with past or ongoing infection with a hepadnavirus. Proc Natl Acad Sci USA 89:3874–3878
Moroy T, Marchio A, Etiemble J, Trepo C, Tiollais P, Buendia MA (1986) Rearrangement and enhanced expression of c-myc in hepatocellular carcinoma of hepatitis virus infected woodchucks. Nature 324:276–279
Ray RB, Lagging LM, Meyer K, Ray R (1996) Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol 70:4438–4443
Acknowledgements
We thank Dr. Charles M. Rice for providing us with the p90/HCVFL-long pU plasmid, and Dr. Li-Yu Wang for the analysis of statistics. This work was supported by grants from the National Science Council of Taiwan (NSC 94-2320-B-320-014 and NSC953112B320001-02) to Dr. Shih-Yen Lo.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below are the electronic supplementary materials.
Supplementary Figure 1
GST-pull down experiment for analysis of the interaction between ARFP/F and MM-1. HCV-1 RNA, which produced the ARFP/F protein with the core protein leader sequence, was synthesized in vitro using the rabbit reticulocyte lysates and labeled with S35-methionine. This frame-shifted ARFP/F protein could be pulled-down by GST-MM-1 (lane 3), but not by the GST control (lane 2). Lane 1, the S35-methionine labeled ARFP/F protein prior to the pull-down experiment. The arrow denotes the ARFP/F protein band. (73.5 KB)
Supplementary Figure 2a
GST-pull down experiments in cultured cells. Panel A, purified GST (lane 1) and GST-MM-1 (lane 2) used in this experiment; Panel B, HuH7 cells were mock transfected (lanes 1, 2 and 3), or transfected with FN (a.a. 1-62 of ARFP/F with the V5 tag ) (lane 4, 5 and 6) or with FC (a.a. 63-151 of ARFP/F with the V5 tag) (lane 7, 8 and 9). 48 hrs after transfection, cell lysates were extracted and incubated with equal amount of purified GST (lanes 2, 5, 8) and GST-MM-1 (lanes 3, 6, 9). Pull-downed samples were analyzed by Western-blotting using the anti-V5 antibody. FN protein bands were marked by the line while FC protein bands were marked by the arrow.(61 KB)
Supplementary Figure 2b
(92 KB)
Supplementary Figure 3
Analysis of the effect of the HCV ARFP/F protein on c-Myc in HuH7 cells. In all the transfection experiments, 0.05 ug pRL-TK was included to serve as an internal control to monitor the transfection efficiency. For the control experiment, the protein expression plasmid was substituted with vector plasmid. Total DNA amount used in each transfection experiment was 1 ug. (A) Transactivation of the reporter expression by c-Myc in a dose-dependent manner. HuH7 cells were transfected with 0.33 ug p4xE SVP-Luc reporter and the indicated amount of the c-Myc expression plasmid. (B) Suppression of the trans-activation activity of c-Myc by MM1. HuH7 cells were co-transfected with 0.33 ug p4xE SVP-Luc reporter, 0.1 ug pEF-cMyc and different amounts of pcDNA3-MM1 as indicated. (C) Suppression of the inhibitory effect of MM1 on c-Myc by the HCV ARFP/F protein. Cells were transfected with 0.33 ug p4xE SVP-Luc reporter, 0.1 ug pEF-cMyc, 0.4 ug pcDNA3-MM1 and different amounts of ARFP/F expression plasmid as indicated. Total DNA amount used in the panel C is 2 ug. (80.5 KB)
Supplementary Figure 4
Enhancement of the c-Myc activity in stable cell lines that expressed the HCV ARFP/F protein. Panel A, the expression of the ARFP/F protein without the core leader sequence (marked by arrow). Stable cells were lysed, immunoprecipitated with rabbit anti-ARFP/F polyclonal antibody and then Western-blotted with the same anti-ARFP/F antibody. Lane 1, parental HuH7 cells; Lanes 2-4, three different stable cell clones that expressed the ARFP/F protein. Panel B, the relative luciferase activit es of parental HuH7 cells and three different stable cell clones that expressed with the truncated ARFP/F protein. Cells were transfected with the reporter and analyzed 72 hrs after transfection for firefly and renilla luciferase activities. The latter served as the control for monitoring the transfection efficiencies. The results shown represent the average of three independent experiments. (77.4 KB)
Rights and permissions
About this article
Cite this article
Ma, HC., Lin, TW., Li, H. et al. Hepatitis C virus ARFP/F protein interacts with cellular MM-1 protein and enhances the gene trans-activation activity of c-Myc. J Biomed Sci 15, 417–425 (2008). https://doi.org/10.1007/s11373-008-9248-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11373-008-9248-9