Abstract
Purpose
Soil microbial communities and related key ecological processes play critical roles in timberline delineation and soil carbon balance in alpine ecosystems, which are highly vulnerable to climate change. Accordingly, understanding their geographical differentiation will facilitate recognition of ecosystem functions and improve soil carbon models. In this study, we explored the biogeographic patterns of soil bacterial communities and their mechanisms in maintaining soil carbon balance in an alpine timberline ecosystem of the Sygera Mountains, Southeast Tibet.
Materials and methods
Soil samples were collected from typical forest belts above and below the timberline. The abundance and composition of bacterial communities, as well as functional genes, were assessed using the gene chip technology. The relationship of key microbial taxa, functional genes, and soil carbon maintenance was investigated using random forest analysis, multi-model inference, and structural equation modeling.
Results and discussion
The shrubland soil bacterial community exhibited greater diversity compared with the coniferous forest community, with higher Shannon Index and more functional genes at the taxonomic and functional levels, respectively. Bacterial community composition differed between the two forest types, with copiotrophic bacteria more abundant in shrubland, and oligotrophic bacteria more abundant in coniferous forest. The shrubland community was also more efficient at utilizing labile organic carbon, while the coniferous forest community utilized recalcitrant organic carbon more efficiently. Genes related to labile carbon degradation were more intense in shrubland, while genes related to recalcitrant carbon degradation were more concentrated in the coniferous forest. Soil temperature and C:N ratio were dominant drivers of bacterial community composition and function. Besides key soil-environment and microbial properties, certain bacterial taxa and functional genes also exerted unique roles in soil carbon variation.
Conclusions
Significant differences exist in soil bacterial community composition and functions between the two forest types above and below the timberline of the Sygera Mountains. These differences may be attributed to soil temperature and soil C:N ratio. Coupling these microbial variables into the earth system model can improve the predictive power of the carbon feedback process in terrestrial ecosystems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Alpine timberlines are important boundaries of ecosystems in altitudinal zonation that are highly vulnerable to climate change and therefore considered ideal monitors of changes in climate (Graumlich et al. 2005). However, delineation of timberlines is challenging because they are characterized by changes in aboveground vegetation communities as well as soil microbial communities, and the involvement of these microbial communities in key ecological processes is important to their definition. Studying geographical differentiation of the microbial community structure and function of timberlines can provide microbial indicators and data that can be used for timberline delineation, as well as contribute to understanding how alpine ecosystems function and respond to global climate change. However, studies of the distribution characteristics, evolutionary processes, and material cycling involved in timberline ecosystems have primarily focused on plant communities (Yang et al. 2008; He et al. 2009; Xu et al. 2010; Liang et al. 2016; Wang et al. 2017; Shen et al. 2017; Zhou et al. 2018; Guo et al. 2019), while limited attention has been given to the overall microbial community structure and function (Thebault et al. 2014; Ding et al. 2015; Shen et al. 2016). Recently, Mayor et al. (2017) compared differences in vegetation, soil nutrients, and microbial community structure and function in timberlines of different elevations around the world and found that global warming may cause disturbances in soil carbon cycling processes and functions in alpine ecosystems.
Soil carbon turnover and the stability of carbon reservoirs in alpine ecosystems have significant impacts on the global carbon source-sink balance. Moreover, the soil carbon balance process is influenced by both biotic and abiotic factors. While soil properties, climate, spatial distance, and vegetation are known to be key factors involved in predicting soil carbon pools, current carbon cycle models with high uncertainty cannot accurately predict changes in these cycles. Identifying other drivers of soil carbon balance is also a major challenge. Some studies have recently incorporated microbial enzymatic processes into terrestrial ecosystem carbon models, which has greatly improved the performance of models predicting soil carbon fluxes (Wieder et al. 2013, 2015; Hararuk et al. 2015). The large, diverse, and ecologically active microbial communities in soil have enormous potential for predicting soil ecological functions such as organic carbon decomposition, mineralization, and stabilization (Hubbell 2005). Therefore, some researchers have proposed coupling microbial community composition and functional genes into terrestrial ecosystem carbon models to improve carbon flux prediction (Trivedi et al. 2016; Wieder et al. 2015). However, few studies have demonstrated a direct correlation between microbial communities and ecosystem functions, including terrestrial ecosystem carbon balance processes (Rocca et al. 2015; Van der Wal et al. 2015; Liu et al. 2018). This has greatly hindered the inclusion of microbial communities in earth system models and other biogeochemical process models.
Southeast Tibet has the world’s highest timberline distribution and a typical timberline landscape, making it an ideal location for studying changes in alpine ecosystems and timberlines. Therefore, this study investigated two typical forest belts located above and below the timberline in the Sygera Mountains of Southeast Tibet. Specifically, the patterns and mechanisms that regulate the composition and function of soil bacterial communities near the timberline were investigated, as was the soil carbon balance driven by microbial communities. The results presented herein provide microbial indicators and evidence for timberline delineation on the Tibet Plateau as well as important information that will be valuable to studying timberline ecosystem functions such as carbon turnover and source-sinks.
The objectives of this study were to (1) describe the differences in composition and diversity of soil bacterial communities between typical forest belts above and below the timberline, (2) describe the differences in carbon and nitrogen cycling functional genes of soil bacterial communities between the two forest belts, (3) determine the dominant environmental drivers that regulate the differences in soil bacterial community composition and function between the two forest belts, and (4) identify key microbial taxa and functional genes that drive soil carbon content in timberline ecosystems based on an empirical model and elucidate their importance and role in the soil carbon balance pathway.
2 Materials and methods
2.1 Soil sampling and laboratory analysis
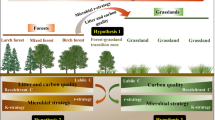
The Sygera Mountain, which is located in the middle and lower reaches of the Yarlung Zangbo River in Southeast Tibet, encompasses an area of approximately 2300 km2. The mountain, which is situated at the boundary between humid and sub-humid climate zones, is influenced by the warm and humid monsoon from the Indian Ocean and characterized by warm winters and cool summers with distinct wet and dry seasons. The average annual temperature is −0.73 °C and the annual precipitation is greater than 800 mm. The area is primarily underlain by acidic brown soil. Because the study area has a large altitude span of 143 to 7728 m above sea level, the mountain is characterized by altitudinal zonation and different climatic zones (subtropical, temperate, and frigid). Farmland is mainly distributed in the low-altitude area below 3500 m, and is mostly used for barley cultivation. As shown in Fig. 1, the alpine temperate dark coniferous forests are distributed between 3300 and 4300 m above sea level, while alpine shrublands and meadows are located at greater than 4300 m above sea level (Li et al. 2015). The main vegetation types in the coniferous forests include A. georgei var. smithii and P. likiangensis var. linzhiensis, while the shrublands and meadows are dominated by alpine rhododendron composed of R. strigillosum var. monosematum, R. tanastylum Balf. F. et K. Ward var. lingzhiense Fang f., and R. corchorifolius L. f.. Overall, Sygera Mountain has a typical alpine timberline landscape. Field sampling on Sygera Mountain was conducted as shown in Fig. 1. Five replicate soil samples were collected from the topsoil (0–15 cm) of two typical forest belts above and below the timberline (4320 m above sea level) and used to evaluate the patterns and control mechanisms of soil microbial communities in the timberline ecosystem. To reduce the likelihood of pseudo-replication, soil replicates were collected using a random procedure from points at least 500 m apart. Each replicate sample is a mixture of three adjacent samples within a 10-m quadrat. Before sampling, all stones, small animals, debris, and litter were removed from the sites. Samples were collected using an AMS split-core sampler (AMS Inc., USA) equipped with a 5-cm-wide and 15-cm-tall probe.
Each soil sample was subsequently divided into subsamples for DNA sequencing and physicochemical soil analyses. Soil DNA extraction and sequencing based on 16S rRNA gene barcoding, selection, and assignment of Operational Taxonomic Units (OTUs) were conducted as described by Yang et al. (2020). The sequence dataset was deposited in the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/) under the accession number SRP076574. The final sample-by-OTU data matrix and annotated taxonomy file were used to calculate the abundance of phyla and diversities. The remaining sub-samples were air-dried and crushed to a particle size of approximately 2 mm for physicochemical analyses.
The soil organic carbon (SOC), total nitrogen, C:N ratio, pH, and soil moisture were measured. For SOC determination, samples were treated with 0.1 mol/L hydrochloric acid to remove inorganic carbon, after which they were dried in a ventilated cabinet, then subjected to dry combustion at 1100 °C using an HT1300 Multi C/N 3100 analyzer (Analytik Jena AG, Germany). The total nitrogen was measured using the Kjeldahl method. The soil C:N ratio was then calculated by dividing the soil organic carbon by the total nitrogen. Soil pH was measured using a suspension of soil and water at a 1:1 ratio. Soil moisture was the absolute water content, which was the water content determined to be in 100 g of dried soil using the oven-drying method. All analyses were conducted as described by Bao (2000).
2.2 GeoChip gene sequencing
The GeoChip is commonly used in metagenomics because of its convenience, speed, accuracy, and stability. This method can detect in situ functional genes and metabolic processes of different microbial communities, including carbon, nitrogen, phosphorus, and sulfur cycles (Zhou et al. 2015). In this study, we followed the protocol established by Yang et al. (2014) to fluorescently label qualified DNA samples, hybridize them onto a chip, detect signals, and preprocess the data. The specific procedures were as follows:
-
1.
Fluorescent labeling: Cy-5 fluorescent dye was employed to label DNA samples using a random primer method. After labeling, the DNA was purified using a QIA kit (Qiagen, Valencia, CA) and then dried at 45 °C for 45 min in a SpeedVac (ThermoSavant, Milford, MA, USA).
-
2.
Chip hybridization and image scanning: After adding 120 µL of hybridization buffer to the labeled DNA samples, sample hybridization was performed at 42 °C for 16 h using the MAUI hybridization platform (BioMicro, Salt Lake City, UT, USA). The hybridized chip was then scanned using a NimbleGen MS200 scanner (Roche, Madison, WI, USA) with a 633 nm laser wavelength, 100% laser intensity, and 75% photomultiplier tube detector gain.
-
3.
Data preprocessing: After scanning, the chip signals were preprocessed using the University of Oklahoma’s microarray data manager (http://ieg.ou.edu/microarray/) to reduce signal differences between samples, decrease random errors, improve the data distribution, and make data more suitable for further analysis and statistics. First, low-quality signals with a signal-to-noise ratio of less than 2.0 were removed. Next, genes that were detected by only one replicate sample were removed so that only individual probes that were detected by two or more replicate samples remained. Finally, the gene chip data were normalized and standardized by calculating the relative signal intensity of the probe. Specifically, the signal intensity of a certain probe in a sample was divided by the total signal intensity of all probes in the sample. This value was then multiplied by the mean total probe signal intensity of all samples, after which the natural logarithm of the relative signal intensity of the probe was taken.
2.3 Data statistics and analysis
Differences in dominant taxon abundance, diversity, and carbon–nitrogen functional genes of soil bacterial communities between typical forest belts above and below the timberline were identified by T-tests after confirming that the data were normally distributed. Differences in bacterial community structure between communities at the taxonomic and functional levels were identified using the community structure dissimilarity test (Adonis) based on the Bray–Curtis dissimilarity distance matrix (Anderson 2001; Bray and Curtis 1957), with the vegan package in R (R Core Team 2014). The relationship between community structure at the taxonomic and functional levels and environmental driving factors was analyzed using the partial Mantel test, redundancy analysis (RDA), and variation partitioning analysis (VPA) with the stats and vegan packages in R.
The importance and pathways of microbes comprising the timberline ecosystem to soil carbon content prediction were investigated using the random forest (RF) method, multi-model inference, and structural equation modeling (SEM).
-
1.
The RF algorithm was used to identify key microbial variables that predict carbon storage, including taxa and functional genes. The variable importance was evaluated based on the increase in mean squared error (MSE) caused by variable permutation, and variables with an MSE increase greater than 3% were selected for further analysis. The RF model was executed using the randomForest package in R.
-
2.
Multi-model inference based on the Second-order Akaika’s Information Criterion (AICc) and linear regression was employed to evaluate the unique effects of microbial variables compared with soil environmental variables on soil carbon prediction. Microbial variables used in this analysis were sorted based on their variable importance determined by the RF algorithm. The accuracy of all models generated by independent prediction variables (soil, environmental, and microbial) was evaluated using AICc, with a smaller AICc value indicating a better model fit. ΔAICc > 2 was used as an indicator to evaluate significant differences between models, and the optimal model was selected accordingly. The multi-model inference method was executed using the glmulti package in R.
-
3.
SEM was used to evaluate the direct and indirect effects of soil, environmental, and microbial variables on soil carbon storage. The model fit accuracy and performance were evaluated using the absolute fit index χ2/df and the root mean square error of approximation (RMSEA), respectively. The SEM was executed in AMOS.
3 Results
3.1 Soil environment and microbial diversity of the timberline ecosystem
The habitat gradient between the two typical forest belts in the timberline ecotone was found to be extremely steep, with high environmental heterogeneity (Table 1). Temperature was a key factor influencing timberline formation, and shrubland soil temperature was significantly higher than that of the coniferous forest (P = 0.001). The SOC content, soil C:N ratio, and soil moisture of the coniferous forest were significantly higher than that of the shrubland, while the soil total nitrogen content of the shrubland was significantly higher than that of the coniferous forest. Therefore, the shrubland had higher soil nutrient levels and better air and water permeability than the coniferous forest.
The Shannon index of soil bacterial community diversity in the shrubland was significantly higher than that in the coniferous forest (P < 0.05). A greater number of functional genes were detected in the shrubland soil bacterial community, resulting in significantly higher functional diversity than the coniferous forest (P < 0.01). Non-parametric tests for community dissimilarity (Adonis) showed significant differences in microbial community composition (F = 9.26, P < 0.01) and functional structure (F = 11.56, P < 0.01) between the upper and lower forest belts in the timberline.
3.2 Microbial composition and carbon–nitrogen cycling functional genes in the timberline ecosystem
Taxonomic annotation based on OTUs revealed 48 phyla. Of these, Acidobacteria (25%), Proteobacteria (21%), Planctomycetes (13%), Verrucomicrobia (8%), Chloroflexi (6%), AD3 (6%), Gemmatimonadetes (5%), Bacteroidetes (4%), Actinobacteria (3%), Nitrospirae (3%), and Firmicutes (2%) were the 11 most abundant phyla, with a cumulative relative abundance of 96%. Proteobacteria included four dominant classes, Alphaproteobacteria (13%), Betaproteobacteria (5%), Gammaproteobacteria (3%), and Deltaproteobacteria (3%). Among these, Alphaproteobacteria and Betaproteobacteria were the two most dominant classes, accounting for 85% of the total Proteobacteria. The relative abundance of Betaproteobacteria, Firmicutes, Verrucomicrobia, and Gemmatimonadetes was significantly higher in the shrubland than in the coniferous forest, while that of Planctomycetes, Chloroflexi, and Acidobacteria was higher in the coniferous forest (Fig. 2).
A total of 120 functional genes involved in soil carbon cycling were detected using GeoChip, including 60 involved in carbon degradation and 57 involved in carbon fixation. There were significant differences in the intensities of 23 carbon degradation functional genes between the coniferous forest and the shrubland (Fig. 3). Genes encoding enzymes for the degradation of labile carbohydrates such as monosaccharides, disaccharides, and starches were generally more intense in the shrubland. Genes encoding enzymes for the degradation of recalcitrant compounds such as lipids and aromatic compounds such as lignin were generally more intense in the coniferous forest.
Among the 57 investigated carbon fixation functional genes, 21 that participate in six natural carbon fixation pathways differed significantly between the coniferous forest and the shrubland (Fig. 3). Specifically, genes encoding enzymes for aerobic carbon fixation pathways such as the Calvin cycle, 3-hydroxypropionate/4-hydroxybutyrate cycle, and 3-hydroxypropionate bicycle were generally more intense in the shrubland, except for the gene encoding ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in the Calvin cycle, which was more intense in the coniferous forest. Genes encoding enzymes for anaerobic carbon fixation pathways such as the dicarboxylate/4-hydroxybutyrate cycle and reductive tricarboxylic acid cycle were generally more intense in the coniferous forest, except for those encoding phosphoenolpyruvate carboxylase (PEPC) and aconitate hydratase (AcnA), which were more intense in the shrubland.
The intensity of aerobic carbon fixation pathway genes was significantly higher in the shrubland, while that of anaerobic carbon fixation pathway genes was significantly higher in the coniferous forest. This was mainly because of significant differences in soil aeration between the two forest types. In the cold fir coniferous forest, the closed canopy significantly reduced the surface water evaporation rate caused by solar radiation, and the thicker litter layer effectively intercepted and retained water, resulting in significantly higher soil moisture and thus reduced soil aeration.
The GeoChip detected a total of 20 functional genes involved in nitrogen cycling processes, six of which showed significant differences in intensity between the coniferous forest and shrubland (Fig. 4). The norB, nosZ, and nifH genes from the coniferous forest had higher intensity than those from the shrubland. These norB and nosZ genes are involved in the reduction of nitric oxide (NO) and nitrous oxide (N2O) to nitrogen (N2), while the nifH gene is involved in nitrogen fixation. Overall, these findings indicate that there is faster nitrogen oxide greenhouse gas conversion and nitrogen fixation in the coniferous forest than in the shrubland. In contrast, the amoA, narG, nasA, and nirA genes from shrubland had higher intensity than those from the forest. These genes are involved in the acceleration of nitrification and denitrification processes. In addition, the intensity of the amo gene from shrubland was also more intense, although this difference was not significant. The presence of this gene has the potential to increase soil nitrate content. Furthermore, genes involved in the reduction of nitrate as an electron acceptor (e.g., the assimilatory nitrate reductase genes nasA and nirA and the denitrification gene narG) showed greater intensity in the shrubland than in the forest.
Relative changes in detected N-cycling genes in the coniferous forest relative to the shrubland. Note: The percentage value for each detected gene indicates the relative fold change. The percentage was calculated by dividing the total signal intensities of each gene by the total signal intensities of all detected N-cycling genes, and then weighted by the fold changes (coniferous forest/shrubland) of each gene. Red- and green-colored genes indicated higher and lower signal intensities in the coniferous forest than in the shrubland, respectively. Grey-colored genes were not detected in samples. ***P < 0.001; **P < 0.01; *P < 0.05
3.3 Environmental factors driving microbial community variation in the timberline ecosystem
RDA biplots were used to investigate the influence of environmental variables on variations in soil bacterial community distribution (Fig. 5). Environmental variables explained a total of 65.2% and 60.7% of the variation in soil bacterial composition and function, respectively. The soil bacterial communities of the two forest types above and below the timberline were well separated along the first two axes of the RDA biplot. Soil temperature, C:N ratio, and normalized difference vegetation index (NDVI) were the environmental variables that had the greatest influence on bacterial community distribution (indicated by the longest arrows).
The partial Mantel test was used to eliminate the interference caused by autocorrelation between environmental factors and to examine the correlations between each environmental factor and the composition and function structure of bacterial communities (Table 2). Soil temperature and soil C:N ratio were found to be most strongly correlated with the composition and function structure of bacterial communities, followed by mean annual temperature, NDVI, and soil moisture. VPA was used to explore the marginal and conditional effects of each environmental variable to compare the contribution of environmental factors to the variation in bacterial communities (Table 3). This analysis showed that soil temperature, C:N ratio, soil moisture, and NDVI were the four most important environmental variables, explaining more than 40% of the variation. Evaluation of the marginal effects revealed that temperature and C:N ratio explained 25.8% and 20.6% of the variation in bacterial community composition, as well as 19.8% and 16.3% of the variation in bacterial community function, respectively, making them the two variables with the highest independent explanatory power. Collectively, these results suggest that soil temperature and C:N ratio were the dominant drivers of spatial variation in the composition and function structure of soil bacterial communities in the timberline of Sygera Mountain.
3.4 Soil carbon content driven by microbes in the timberline ecosystem
SOC content exhibited significant differences in the forest belts above and below the timberline, with a significantly higher SOC content being observed in the coniferous forest than that in the shrubland (Table 1). SOC content was strongly correlated with soil environmental variables such as soil C:N ratio, mean annual temperature (MAT), NDVI, and slope (Table 4). These soil and environmental attributes also showed significant differences between the two forest belts (Table 1).
Using a random forest model, we identified key microbial taxa and carbon cycling functional genes that predicted the SOC content (Fig. 6). These taxa included copiotrophic bacteria such as Betaproteobacteria and Bacteroidetes, as well as oligotrophic bacteria such as Planctomycetes and Chloroflexi. The functional genes identified included fungal glucose oxidase genes (Glucose_oxidase_fungi) and fungal alpha-galactosidase genes (Alpha_galactosidase_fungi) involved in the degradation of monosaccharides and disaccharides (glucose and lactose). Other functional genes included manganese peroxidase genes (mnp) and ligninase genes involved in lignin degradation, mannanase genes involved in hemicellulose degradation, 3-hydroxypropionate/4-hydroxybutyrate cycle genes (3HP_CoAs) involved in carbon fixation, and propionyl-CoA synthetase genes (pcs) involved in the 3-hydroxypropionate cycle. Furthermore, the functional diversity index (FDI) was highly correlated with SOC (Table 4).
An optimal linear model based on multi-model inference explained 94% of the variation in SOC. The prediction variables included soil environmental properties (C:N ratio and NDVI), microbial composition (Betaproteobacteria and Planctomycetes), and carbon cycling functional genes (Glucose_oxidase_fungi and mnp) (Table 5). The results showed that, when microbial composition and functional genes were removed from the optimal model, the AICc value increased and the explanatory power of the model decreased (R2 = 0.87, ΔAICc = 13.49). Therefore, microbial variables could partially explain the variation in SOC and had unique effects that could not be explained by soil environmental variables.
A structural equation model was used to analyze the direct and indirect effects of the timberline ecosystem, soil environmental properties, and microbes on SOC content so that the mechanisms driving organic carbon content could be understood from a system level. When the optimal model based on multi-model inference was used to parameterize and standardize the structural equation model, the structural equation model explained 90% of the SOC variation (R2 = 0.90, Fig. 7). Additionally, the impact paths of the driving factors in the model reached a significant level (P < 0.05), and χ2/df = 1.343 (P = 0.25) and RMSEA = 0.09 (P = 0.23) met the evaluation criteria, indicating that the model was reasonable.
Based on the path coefficients of the model (Fig. 7, Table 6), soil properties, microbial composition, and functional genes had a strong direct impact on SOC content, with direct effects of 0.72, 0.27, and −0.12, respectively. The direct effect of the timberline ecosystem was only 0.10 because it mainly indirectly affected the soil carbon content by influencing vegetation, soil properties, microbial composition, and functional genes. The direct effect of soil C:N ratio on carbon content was the most significant, while it indirectly affected carbon content by regulating microbial composition. Overall, the total effect of the soil C:N ratio was 0.80, making it the main non-biological factor affecting the soil carbon content in timberline ecosystems. The direct effect of microbial composition was 0.27, while its indirect effect on soil carbon content through effects on carbon cycling functional genes was −0.02. Overall, the total effect of microbial composition was 0.25, making it the main microbial factor affecting soil carbon content in timberline ecosystems. In addition, functional genes encoding microbial monosaccharide and lignin-degrading enzymes had a direct effect of −0.12, indicating they were an important microbial factor affecting soil carbon content in timberline ecosystems.
4 Discussion
4.1 Dominant environmental factors driving variation in timberline bacterial communities
In this study, soil temperature and soil C:N ratio were strongly correlated with the bacterial community structure at both taxonomic and functional levels, which were identified as the dominant environmental factors driving variation in soil microbial communities in timberline ecosystems. Bacterial taxonomic and functional diversity in the soil of the sparse shrubland above the timberline were significantly higher than those in the closed coniferous forest below the timberline, which was consistent with the results of previous studies (Ding et al. 2015; Shen et al. 2016; Yuan et al. 2015). In another study, temperature was found to be the dominant environmental factor driving formation of the timberline (Harsch et al. 2009). The changes in vegetation and microclimate and soil conditions induced by temperature fluctuations significantly affected the structure and function of soil microbial communities above and below the timberline (Harsch et al. 2009). The soil temperature in the shrubland was significantly higher than that in the coniferous forest. This finding was consistent with the results of previous studies (Ding et al. 2015; Yuan et al. 2015) that showed closed canopies reduce the amount of solar radiation received by the soil, resulting in significantly lower soil temperatures in closed forests in timberline ecosystems than in shrub areas at higher altitudes. Temperature can affect the metabolic rate of microbes on a kinetic level, thereby affecting growth and diversity. As environmental temperature increases, taxon richness increases rapidly (Allen et al. 2002; Davidson and Janssens 2006; Hendershot et al. 2017). For example, Nottingham et al. (2018) found that microbial diversity along an altitude gradient decreased with decreasing temperature under relatively constant soil conditions. However, temperature may also indirectly affect microbes by influencing plant growth and soil properties (Mitchell et al. 2012; Yuan et al. 2015). For instance, Wang et al. (2015) reported that the strong climatic changes along an altitudinal gradient led to vertical differentiation of vegetation, resulting in higher soil C:N ratios and lower bacterial community diversity in higher altitude areas. In the present study, there were significant differences in vegetation composition above and below the timberline, with the coniferous forest below the timberline being mainly composed of cold fir and the shrubland above the timberline mainly composed of rhododendron and Rubus. Compared with the shrubland, the coniferous forest has a higher leaf litter C:N ratio, which led to higher soil C:N ratios and lower soil nutrient availability, limiting microbial growth and activity (Djukic et al. 2010; Zheng et al. 2018). However, alpine shrub vegetation can adapt to and buffer extreme low temperatures and improve the biochemical activity and microsite survival conditions of soil, making it more conducive to microbial growth (Zhou and Ma 2013; He et al. 2014).
4.2 Differences in taxa and carbon cycling functions of timberline bacterial communities
Similarly, the differences in taxonomic abundance and carbon cycling functional genes between forest belts above and below the timberline were closely related to soil nutrient status. At the taxonomic level, soil bacteria can be divided into copiotrophic bacteria and oligotrophic bacteria based on their carbon mineralization potential and growth rate (Fierer et al. 2007). Copiotrophic bacteria, which prefer nutrient-rich environments and primarily decompose labile soil carbon pools, include Bacteroidetes, Alphaproteobacteria, and Betaproteobacteria. Oligotrophic bacteria, which mainly decompose stable or recalcitrant soil carbon pools, include Acidobacteria, Planctomycetes, and Chloroflexi (Fierer et al. 2007; Bastian et al. 2009; Ramirez et al. 2012). In the present study, some bacterial taxa such as Betaproteobacteria and Verrucomicrobia were more abundant in the nutrient-rich shrubland, while Planctomycetes and Chloroflexi were more abundant in the nutrient-poor coniferous forest.
At the functional level, the intensity of genes encoding enzymes that degrade labile organic carbon was higher in the shrubland, while the intensity of genes encoding enzymes that degrade recalcitrant or stable organic carbon was higher in the coniferous forest. Therefore, the cold fir coniferous forest was more efficient than shrubland at degradation and utilization of recalcitrant organic carbon. Other studies of Sygera Mountain also showed that the coniferous forest had higher levels of stable organic carbon in the soil and lower levels of active organic carbon when compared with shrubland (Ma et al. 2013; Zhou et al. 2015). The litter of cold fir coniferous forests is harder and contains more recalcitrant substances, particularly lignin (Zhong and Xin 2004; Zheng et al. 2018). As a result, long-term environmental selection has led to changes in bacterial community composition and organic carbon degradation functions. Recalcitrant and stable organic carbon can persist in soil carbon pools, where it plays an important role in maintaining the long-term stability of soil carbon pools (Purton et al. 2015; Zhang et al. 2018). With the timberline constantly moving upward under the background of global warming, the degradation and utilization of stable organic carbon will accelerate. This could break the soil carbon source-sink balance of the timberline and accelerate the positive feedback effect of climate warming.
4.3 Soil carbon balance driven by taxa and functional genes in timberline ecosystems
Studies of Sygera Mountain have indicated a high correlation between vegetation status, soil C:N ratio, and SOC content (Ma et al. 2012), which is consistent with the results of our study. The optimal model identified for predicting organic carbon in the present study suggested that microbial factors including Betaproteobacteria and Planctomycetes, fungal functional genes encoding glucose oxidase (Glucose_oxidase_fungi), and lignin-degrading enzyme genes (mnp) had unique effects on soil carbon variation in the timberline ecosystem in addition to the effects of the NDVI and soil C:N ratio. We also used structural equation modeling to evaluate the roles of soil environmental factors and microbial factors in driving soil carbon balance at the system level. The soil C:N ratio directly and indirectly affected SOC content and was the most important non-biological factor driving soil carbon content. Previous studies have shown that the soil C:N ratio is a key factor controlling soil carbon mineralization rate, and an excessively high soil C:N ratio suppresses microbial decomposition of organic matter (Djukic et al. 2010). Additionally, the soil C:N ratio can have indirect effects on carbon content by affecting microbial community composition. Numerous studies have shown that altering the soil C:N ratio significantly changes the structure and diversity of microbial communities, thereby changing the diversity and specificity of carbon source utilization in the community (Tamilselvi et al. 2015; Xing et al. 2016). The direct effects of microbial composition were second only to the soil C:N ratio; therefore, these effects were the most important biological driving factor. As the primary biological agent of soil carbon decomposition and loss, microbes play an important role in soil carbon balance (Trivedi et al. 2016). Specifically, copiotrophic bacteria (e.g., Betaproteobacteria) decompose the labile carbon pool and grow rapidly, while oligotrophic bacteria (e.g., Planctomycetes) decompose the stable carbon pool. In addition, functional genes encoding carbon-degrading enzymes, such as genes encoding glucose oxidase (Glucose_oxidase_fungi), which degrade monosaccharides, and genes encoding manganese peroxidase (mnp), which degrade lignin, were found to have direct effects on soil carbon content. Therefore, these genes are also important biological factors driving carbon content. Monosaccharides are an important component of active carbon sources that have strong microbial activity and directly participate in soil biochemical transformation processes; therefore, their decomposition rate directly affects the capacity of the active carbon pool (Coleman et al. 1983). Lignin is an important component of the stable carbon pool that directly determines the accumulation and long-term carbon sequestration capacity of stable soil carbon pools through the speed of microbial degradation (Peng et al. 2004; Purton et al. 2015).
Microbial community composition and functional genes play a unique role in explaining variations in soil carbon in timberline ecosystems, with unique effects that could not be explained by other environmental variables. These results demonstrate the importance of microbial communities in maintaining soil ecosystem function and stability. Incorporating these key microbial variables into earth system models may improve the predictive ability of land ecosystem carbon feedback processes. As the timberline continues to shift upward under climate change, changes in microbial community structure and function will inevitably affect and disrupt soil carbon source-sink balance in timberline ecosystems. Our results provide theoretical support for the management of timberline soil carbon through artificial management measures to change soil environmental conditions and optimize community structure, thereby improving the soil carbon sink potential of timberline ecosystems.
5 Conclusions
This study revealed significant differences in soil bacterial community structure and function between two forest types (shrubland and coniferous forests) above and below the timberline of Sygera Mountain in the Southeast Tibet Plateau. Soil temperature and soil C:N ratio emerged as dominant driving factors which were responsible for these significant differences. The nutrient-rich shrubland demonstrated a higher soil bacterial community diversity than the coniferous forest, with a higher abundance of copiotrophic bacteria and lower abundance of oligotrophic bacteria at the taxonomic level. Similarly, at the functional level, the intensity of genes encoding the degradation of labile organic carbon was higher in the shrubland, while genes encoding the degradation of recalcitrant organic carbon were higher in the coniferous forest. Therefore, the shrubland was more efficient at decomposing and utilizing labile organic carbon, while the coniferous forest was more efficient at decomposing recalcitrant organic carbon. The study also established that the key bacterial taxa and functional genes involved in carbon degradation in the timberline ecosystem were important biological factors driving the timberline soil carbon balance, and that they had a unique explanatory effect on variations in timberline soil carbon. In the context of global climate change, changes in the structure and function of microbial communities will inevitably affect and disrupt the soil carbon source-sink balance in timberline ecosystems. Overall, this study provides a theoretical basis that will be useful for the management of timberline soil carbon source-sinks on the Tibet Plateau.
Data availability
The data used for the analyses presented in this work is available from the corresponding author on reasonable request.
References
Allen AP, Brown JH, Gillooly JF (2002) Global biodiversity, biochemical kinetics, and the energetic-equivalence rule. Science 297:1545–1548. https://doi.org/10.1126/science.1072380
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. https://doi.org/10.1111/j.1442-9993.2001.01070.pp.x
Bao S (2000) Soil and agricultural chemistry analysis. Chinese Agriculture Publication, Beijing
Bastian F, Bouziri L, Nicolardot B, Ranjard L (2009) Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol Biochem 41:262–275. https://doi.org/10.1016/j.soilbio.2008.10.024
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:326–349. https://doi.org/10.2307/1942268
Coleman DC, Reid C, Cole C (1983) Biological strategies of nutrient cycling in soil systems. Adv Ecol Res 13:1–55. https://doi.org/10.1016/S0065-2504(08)60107-5
Davidson EA, Janssens IA (2006) Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440:165–173. https://doi.org/10.1038/nature04514
Ding J, Zhang Y, Deng Y, Cong J, Lu H, Sun X, Yang C, Yuan T, Van Nostrand JD, Li D (2015) Integrated metagenomics and network analysis of soil microbial community of the forest timberline. Sci Rep 5:1–10. https://doi.org/10.1038/srep07994
Djukic I, Zehetner F, Mentler A, Gerzabek MH (2010) Microbial community composition and activity in different Alpine vegetation zones. Soil Biol Biochem 42:155–161. https://doi.org/10.1016/j.soilbio.2009.10.006
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. https://doi.org/10.1890/05-1839
Graumlich LJ, Waggoner LA, Bunn AG (2005) Detecting global change at alpine treeline: coupling paleoecology with contemporary studies. Global change and mountain regions: an overview of current knowledge. Springer, Dordrech. https://doi.org/10.1007/1-4020-3508-X_50
Guo B, Wang X, Zhang Y (2019) Effects of accumulated and threshold temperatures on the radial growth of Abies faxoniana in the alpine timberline, Western Sichuan Plateau. Acta Ecol Sin 39:895–904. https://doi.org/10.5846/stxb201803300686
Hararuk O, Smith MJ, Luo Y (2015) Microbial models with data-driven parameters predict stronger soil carbon responses to climate change. Global Change Biol 21:2439–2453. https://doi.org/10.1111/gcb.12827
Harsch MA, Hulme PE, McGlone MS, Duncan RP (2009) Are treelines advancing? A global meta-analysis of treeline response to climate warming. Ecol Lett 12:1040–1049. https://doi.org/10.1111/j.1461-0248.2009.01355.x
He J, Luo T, Xu Y (2009) Characteristics of eco-climate at smith fir timberline in the Sergyemla Mountains, Southeast Tibetan Plateau. Acta Ecol Sin 29:37–46. https://doi.org/10.13287/j.1001-9332.2004.0038
He Y, Xu X, Kueffer C, Zhang X, Shi P (2014) Leaf litter of a dominant cushion plant shifts nitrogen mineralization to immobilization at high but not low temperature in an alpine meadow. Plant Soil 383:415–426
Hendershot JN, Read QD, Henning JA, Sanders NJ, Classen AT (2017) Consistently inconsistent drivers of microbial diversity and abundance at macroecological scales. Ecology 98:1757–1763. https://doi.org/10.1002/ecy.1829
Hubbell SP (2005) Neutral theory in community ecology and the hypothesis of functional equivalence. Funct Ecol 19:166–172. https://doi.org/10.1111/j.0269-8463.2005.00965.x
Li S, Shi Z, Chen S, Ji W, Zhou L, Yu W, Webster R (2015) In situ measurements of organic carbon in soil profiles using vis-NIR spectroscopy on the Qinghai-Tibet plateau. Environ Sci Technol 49:4980–4987. https://doi.org/10.1021/es504272x
Liang E, Wang Y, Piao S, Lu X, Camarero JJ, Zhu H, Zhu L, Ellison AM, Ciais P, Peñuelas J (2016) Species interactions slow warming-induced upward shifts of treelines on the Tibetan Plateau. Proc Natl Acad Sci 113:4380–4385. https://doi.org/10.1073/pnas.1520582113
Liu Y, Delgado-Baquerizo M, Wang J, Hu H, Yang Z, He J (2018) New insights into the role of microbial community composition in driving soil respiration rates. Soil Biol Biochem 118:35–41. https://doi.org/10.1016/j.soilbio.2017.12.003
Ma H, Guo Q, Liu H, Qian D (2013) Soil organic carbon pool at the western side of the sygera mountains, southeast Tibet, China. Acta Ecol Sin 33:3122–3128. https://doi.org/10.5846/stxb201202170210
Ma H, Guo Q, Liu H, Qian D (2012) Variations of soil microbial biomass and readily oxidizable organic carbon along elevation gradient in Sejila Mountain, Tibet. J Soil Water Conserv 26:163–166. https://doi.org/10.13870/j.cnki.stbcxb.2012.04.035
Mayor JR, Sanders NJ, Classen AT, Bardgett RD, Clement J-C, Fajardo A, Lavorel S, Sundqvist MK, Bahn M, Chisholm C (2017) Elevation alters ecosystem properties across temperate treelines globally. Nature 542:91–95. https://doi.org/10.1038/nature21027
Mitchell RJ, Hester AJ, Campbell CD, Chapman SJ, Cameron CM, Hewison RL, Potts JM (2012) Explaining the variation in the soil microbial community: do vegetation composition and soil chemistry explain the same or different parts of the microbial variation? Plant Soil 351:355–362. https://doi.org/10.1007/s11104-011-0968-7
Nottingham AT, Fierer N, Turner BL, Whitaker J, Ostle NJ, McNamara NP, Bardgett RD, Leff JW, Salinas N, Silman MR (2018) Microbes follow Humboldt: temperature drives plant and soil microbial diversity patterns from the Amazon to the Andes. Ecology 99:2455–2466. https://doi.org/10.1002/ecy.2482
Peng X, Zhang B, Zhao Q (2004) A review on relationship between soil organic carbon pools and soil structure stability. Acta Pedol Sin 41:618–623. https://doi.org/10.3321/j.issn:0564-3929.2004.04.019
Purton K, Pennock D, Leinweber P, Walley F (2015) Will changes in climate and land use affect soil organic matter composition? Evidence from an ecotonal climosequence. Geoderma 253:48–60. https://doi.org/10.1016/j.geoderma.2015.04.007
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Ramirez KS, Craine JM, Fierer N (2012) Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Global Change Biol 18:1918–1927. https://doi.org/10.1111/j.1365-2486.2012.02639.x
Rocca JD, Hall EK, Lennon JT, Evans SE, Waldrop MP, Cotner JB, Nemergut DR, Graham EB, Wallenstein MD (2015) Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J 9:1693–1699. https://doi.org/10.1038/ismej.2014.252
Shen C, Shi Y, Ni Y, Deng Y, Van Nostrand JD, He Z, Zhou J, Chu H (2016) Dramatic increases of soil microbial functional gene diversity at the treeline ecotone of Changbai Mountain. Front Microbiol 7:1184. https://doi.org/10.3389/fmicb.2016.01184
Shen W, Zhang L, Luo T (2017) Advances in the study of the limitations of seedling recruitment for alpine timberline forests. Acta Ecol Sin 37. https://doi.org/10.5846/stxb201601230158
Tamilselvi S, Chinnadurai C, Ilamurugu K, Arulmozhiselvan K, Balachandar D (2015) Effect of long-term nutrient managements on biological and biochemical properties of semi-arid tropical Alfisol during maize crop development stages. Ecol Indicators 48:76–87. https://doi.org/10.1016/j.ecolind.2014.08.001
Thébault A, Clément JC, Ibanez S, Roy J, Geremia RA, Pérez CA, Buttler A, Estienne Y, Lavorel S (2014) Nitrogen limitation and microbial diversity at the treeline. Oikos 123:729–740. https://doi.org/10.1111/j.1600-0706.2013.00860.x
Trivedi P, Delgado-Baquerizo M, Trivedi C, Hu H, Anderson IC, Jeffries TC, Zhou J, Singh BK (2016) Microbial regulation of the soil carbon cycle: evidence from gene–enzyme relationships. ISME J 10:2593–2604. https://doi.org/10.1038/ismej.2016.65
van der Wal A, Ottosson E, De Boer W (2015) Neglected role of fungal community composition in explaining variation in wood decay rates. Ecology 96:124–133. https://doi.org/10.1890/14-0242.1
Wang J, Cao P, Hu H, Li J, Han L, Zhang L, Zheng Y, He J (2015) Altitudinal distribution patterns of soil bacterial and archaeal communities along Mt. Shegyla on the Tibetan Plateau. Microb Ecol 69:135–145. https://doi.org/10.1007/s00248-014-0465-7
Wang Y, Liang E, Lu X, Zhu H, Piao S, Zhu L (2017) Are treelines advancing in response to climate warming on the Tibetan Plateau? Chin J Nat 39:179–183. https://doi.org/10.3969/j.issn.0253-9608.2017.03.004
Wieder WR, Bonan GB, Allison SD (2013) Global soil carbon projections are improved by modelling microbial processes. Nat Clim Change 3:909–912. https://doi.org/10.1038/nclimate1951
Wieder WR, Allison SD, Davidson EA, Georgiou K, Hararuk O, He Y, Hopkins F, Luo Y, Smith MJ, Sulman B (2015) Explicitly representing soil microbial processes in Earth system models. Global Biogeochem Cycles 29:1782–1800. https://doi.org/10.1002/2015GB005188
Xing P, Wu X, Gao S, Li H, Zhao T, Zhou X, Shen D, Sun J (2016) Effects of different fertilization on soil microbial community and functional diversity in maize-wheat crop rotation. J Microbiol 36:22–29. https://doi.org/10.3969/j.issn.1005-7021.2016.01.005
Xu Z, Hu T, Zhang L, Zhang Y, Xian J, Wang K (2010) Short-term gas exchange responses of Betula utilis to simulated global warming in a timberline ecotone, eastern Tibetan Plateau, China. Chin J Plant Ecol 34:263–270. https://doi.org/10.3773/j.issn.1005-264x.2010.03.003
Yang Y, Gao Y, Wang S, Xu D, Yu H, Wu L, Lin Q, Hu Y, Li X, He Z (2014) The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J 8:430–440. https://doi.org/10.1038/ismej.2013.146
Yang YY, Zhou Y, Shi Z, Viscarra Rossel RA, Liang ZZ, Wang HZ, Zhou LQ, Yu W (2020) Interactive effects of elevation and land use on soil bacterial communities in the Tibetan Plateau. Pedosphere 30(6):817–831
Yang X, Cui G, Ren Q, Wang J, Huang Y (2008) Plant diversity pattern and stability of the timberline community on Sejila Mountain in Tibet, western China. J Beijing For Univ 30:14–20. https://doi.org/10.13332/j.1000-1522.2008.01.015
Yuan Y, Si G, Wang J, Han C, Zhang G (2015) Effects of microclimate on soil bacterial communities across two contrasting timberline ecotones in southeast Tibet. Eur J Soil Sci 66:1033–1043. https://doi.org/10.1111/ejss.12292
Zhang Z, Li M, Song X, Xue Z, Lv X, Jiang M, Wu H, Wang X (2018) Effects of climate change on molecular structure and stability of soil carbon pool: a general review. Acta Pedol Sin 55:273–282. https://doi.org/10.1016/j.geoderma.2019.07.008
Zheng H, Chen Y, Liu Y, Zhang J, Yang W, Yang L, Li H, Wang L, Wu F, Guo L (2018) Litter quality drives the differentiation of microbial communities in the litter horizon across an alpine treeline ecotone in the eastern Tibetan Plateau. Sci Rep 8:1–11. https://doi.org/10.1038/s41598-018-28150-1
Zhong G, Xin X (2004) Chemical properties of litter in dark coniferous forest of Sejila Mountains in Tibet. Chin J Appl Ecol 15:167–169. https://doi.org/10.13287/j.1001-9332.2004.0038
Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L (2015) High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. Mbio 6:e02288–e2314. https://doi.org/10.1128/mBio.02288-14
Zhou C, Ma H (2013) Distribution of labile organic in soil as affected by vegetation typical of Sygera Mountains, Tibet, China. Acta Pedol Sin 50:1246–1251. https://doi.org/10.11766/trxb201301090018
Zhou T, Narayan PG, Liao L, Zheng L, Wang J, Sun J, Wei Y, Xie Y, Wu Y (2018) Spatio-temporal dynamics of two alpine treeline ecotones and ecological characteristics of their dominate species at the eastern margin of Qinghai-Xizang Plateau. Chin J Plant Ecol 42:1082–1093. https://doi.org/10.17521/cjpe.2018.0082
Funding
This study was supported by the National Natural Science Foundation of China (No.42101061, No.41930754, No.42001047).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Responsible editor: Jizheng He
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, Y., Chen, Q., Zhou, Y. et al. Soil bacterial community composition and function play roles in soil carbon balance in alpine timberline ecosystems. J Soils Sediments 24, 323–336 (2024). https://doi.org/10.1007/s11368-023-03627-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-023-03627-3