Abstract
Purpose
This study was conducted to investigate shifts in the bacterial community structure and soil enzyme activities during long-term reclamation of different land use types in a freshwater wetland.
Materials and methods
Illumina 16S rRNA gene amplification and sequencing and soil extracellular enzymes were used to evaluate changes in bacterial community structure and potential function under four different land use types after 38 years of reclamation: abandoned wetland (AW), vegetable field (VF), pond sediment (PS), rice field (RF) and natural wetland (NW).
Results and discussion
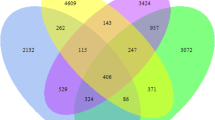
Although bacterial rRNA gene alpha diversity remained unchanged, the bacterial community composition changed significantly under different land uses according to hydrological conditions. Under drying-wetting cycle conditions, RF harbored a similar bacterial community composition as NW, with Acidobacteriales, Holophagae, Planctomycetes, and Spirochaetae being dominant in both. Additionally, AW and VF as upland soil had significantly higher abundance of Gemmatimonadates and Actinobacteria. The composition of permanently flooded soil (PS) had unique features and significantly enriched levels of Chlorobi. The shifts in bacterial community structure were mainly driven by soil moisture content, as well as total organic C and N. However, variations in potential enzyme activities were better explained by soil properties than microbial community composition, suggesting high plasticity of the resident microbial community to environmental conditions.
Conclusions
Rice field harbored a similar bacterial community structure as natural wetland, while the responses of enzyme activities for all land use types differed significantly. The distributions of bacterial community might be influenced by hydrology, while extracellular enzyme activities were closely related to soil properties. Our results suggest that the function of the active microbial community should also be considered in environmental risk models as indicators of ecosystem conversion caused by land use and anthropogenic activity.






Similar content being viewed by others
References
Allison SD (2005) Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol Lett 8:626–635
Argiroff WA, Zak DR, Lanser CM, Wiley MJ (2017) Microbial community functional potential and composition are shaped by hydrologic connectivity in riverine floodplain soils. Microb Ecol 73:1–15
Baldrian P (2014) Distribution of extracellular enzymes in soils: spatial heterogeneity and determining factors at various scales. Soil Sci Soc Am J 78:11
Banerjee S, Bora S, Thrall PH, Richardson AE (2016) Soil C and N as causal factors of spatial variation in extracellular enzyme activity across grassland-woodland ecotones. Appl Soil Ecol 105:1–8
Bang-Andreasen T, Nielsen JT, Voriskova J (2017) Wood ash induced pH changes strongly affect soil bacterial numbers and community composition. Front Microbiol 8:1400
Bastida F, Torres IF, Moreno JL, Baldrian P, Ondoño S, Ruiz-Navarro A, Hernández T, Richnow HH, Starke R, García C (2016) The active microbial diversity drives ecosystem multifunctionality and is physiologically related to carbon-availability in Mediterranean semiarid soils. Mol Ecol 25:4660–4673
Bastida F, Torres IF, Andrésabellán M, Baldrian P, Lópezmondéjar R, Větrovský T, Richnow HH, Starke R, Ondoño S, García C (2017) Differential sensitivity of total and active soil microbial communities to drought and forest management. Glob Chang Biol 23:4185–4203
Bissett A, Richardson AE, Baker G, Thrall PH (2011) Long-term land use effects on soil microbial community structure and function. Appl Soil Ecol 51:66–78
Bowles TM, Acosta-Martínez V, Calderón F, Jackson LE (2014) Soil enzyme activities, microbial communities, and carbon and nitrogen availability in organic agroecosystems across an intensively-managed agricultural landscape. Soil Biol Biochem 68:252–262
Bridgham SD, Cadillo-Quiroz H, Keller JK, Zhuang Q (2013) Methane emissions from wetlands: biogeochemical, microbial, and modeling perspectives from local to global scales. Glob Chang Biol 19:1325–1346
Bu NS, Qu JF, Li G, Zhao B, Zhang RJ, Fang CM (2015) Reclamation of coastal salt marshes promoted carbon loss from previously-sequestered soil carbon pool. Ecol Eng 81:335–339
Burns RG (1982) Enzyme activity in soil: location and a possible role in microbial ecology. Soil Biol Biochem 14:423–427
Canbolat MY, Bilen S, Çakmakçı R, Şahin F, Aydın A (2006) Effect of plant growth-promoting bacteria and soil compaction on barley seedling growth, nutrient uptake, soil properties and rhizosphere microflora. Biol Fertil Soil 42:350–357
Cao XY, Yong-Mei LI, Zhang HZ, Zhang WL, Wang ZL (2010) Differences of phosphorus loss on different soil texture in-situ under simulated rainfall conditions of Dianchi Lake Watershed. J Soil Water Conserv 24:13–17
Chen Y, Yu D, Wang Y, Wu Z, Xie S, Liu Y (2016) Distribution of bacterial communities across plateau freshwater lake and upslope soils. J Environ Sci 43:61–69
Costanza R (2006) Nature: ecosystems without commodifying them. Nature 443:749 author reply 750
Debruyn JM, Nixon LT, Fawaz MN, Johnson AM, Radosevich M (2011) Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl Environ Microbiol 77:6295–6300
Delgado-Baquerizo M, Maestre FT, Reich PB (2016) Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun 7:10541
Ding LJ, Su JQ, Li H, Zhu YG, Cao ZH (2017) Bacterial succession along a long-term chronosequence of paddy soil in the Yangtze River Delta, China. Soil Biol Biochem 104:59–67
Dolinar N, Šraj N, Gaberščik A (2010) Water regime changes and the function of an intermittent wetland. Springer, Netherlands, pp 251–262
Duan Y, Wu F, Wang W, He D, Gu JD, Feng H, Chen T, Liu G, An L (2017) The microbial community characteristics of ancient painted sculptures in Maijishan Grottoes, China. PLoS One 12:e0179718
Eivazi F, Tabatabai MA (1977) Phosphatases in soils. Soil Biol Biochem 9:167–172
Esch EH, Lipson D, Cleland EE (2017) Direct and indirect effects of shifting rainfall on soil microbial respiration and enzyme activity in a semi-arid system. Plant Soil 411:333–346
Fawaz, Naomi M (2013) Revealing the ecological role of Gemmatimonadetes through cultivation and molecular analysis of agricultural soils. Master’s theses, University of Tennessee. https://trace.tennessee.edu/utk_gradthes/1652
Fichtner A, Oheimb GV, Härdtle W, Wilken C, Gutknecht JLM (2014) Effects of anthropogenic disturbances on soil microbial communities in oak forests persist for more than 100 years. Soil Biol Biochem 70:79–87
Gao B, Gupta RS (2005) Conserved indels in protein sequences that are characteristic of the phylum Actinobacteria. Int J Syst Evol Microbiol 55:2401–2412
Garcíamantrana I, Selmaroyo M, Alcántara Baena C, Collado MC (2018) Shifts on gut microbiota associated to mediterranean diet adherence and specific dietary intakes on general adult population. Front Microbiol 9:890
García-Ruiz R, Ochoa V, Hinojosa MB, Carreira JA (2008) Suitability of enzyme activities for the monitoring of soil quality improvement in organic agricultural systems. Soil Biol Biochem 40:2137–2145
Gramss G, Voigt KD, Kirsche B (1999) Oxido-reductase enzymes librated by plant roots and their effects on soil humic material. Chemosphere 38:1481–1494
He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, Huang Z, Wu W, Gu B, Jardine P (2007) GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J 1:67–77
Hua J, Feng Y, Jiang Q, Bao X, Yin Y (2017) Shift of bacterial community structure along different coastal reclamation histories in Jiangsu, Eastern China. Sci Rep 7:10096
Ismail MH, Latiffah H, Hj SZ, Chandrawathani P, Zunita Z, Saleha AA (2016) Physicochemical properties influencing presence of Burkholderia pseudomalleiin soil from small ruminant farms in Peninsular Malaysia. PLoS One 11:e0162348
Ivanova AA, Philippov DA, Kulichevskaya IS, Dedysh SN (2017) Distinct diversity patterns of Planctomycetes associated with the freshwater macrophyte Nuphar lutea (L.) Smith. Antonie Van Leeuwenhoek, pp 1–13
Ji B, Hu H, Zhao Y, Mu X, Liu K, Li C (2014) Effects of deep tillage and straw returning on soil microorganism and enzyme activities. Sci World J 2014:451493
Jiang XT, Peng X, Deng GH, Sheng HF, Wang Y, Zhou HW, Tam FY (2013) Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb Ecol 66:96–104
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3:442–453
Kandeler E, Stemmer M, Klimanek EM (1999) Response of soil microbial biomass, urease and xylanase within particle size fractions to long-term soil management. Soil Biol Biochem 31:261–273
Khan MI, Hwang HY, Kim GW, Kim PJ, Das S (2018) Microbial responses to temperature sensitivity of soil respiration in a dry fallow cover cropping and submerged rice mono-cropping system. Appl Soil Ecol 128:98–108
Knittel K, Boetius A, Lemke A, Eilers H, Lochte K, Pfannkuche O, Linke P, Amann R (2003) Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia Margin, Oregon). Geomicrobiol J 20:269–294
Kumar KV, Lall C, Raj RV, Vedhagiri K, Vijayachari P (2016) Molecular detection of pathogenic leptospiral protein encoding gene (lipL32) in environmental aquatic biofilms. Lett Appl Microbiol 62:311–315
Li Z, Jin Z, Li Q (2017) Changes in land use and their effects on soil properties in Huixian Karst Wetland System. Pol J Environ Stud 26:699–707
Li N, Shao T, Zhu T, Long X, Gao X, Liu Z, Shao H, Rengel Z (2018) Vegetation succession influences soil carbon sequestration in coastal alkali-saline soils in southeast China. Sci Rep 8:9728
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272–1277
Lv W, Liu Z, Yang Y, Huang Y, Fan B, Jiang Q, Zhao Y (2016) Loss and self-restoration of macrobenthic diversity in reclamation habitats of estuarine islands in Yangtze Estuary, China. Mar Pollut Bull 103:128–136
Ma Y, Liu F, Kong Z, Yin J, Kou W, Wu L, Ge G (2016) The distribution pattern of sediment archaea community of the Poyang Lake, the Largest Freshwater Lake in China. Archaea 2016:1–12
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Martínez AT, Speranza M, Ruizdueñas FJ, Ferreira P, Camarero S, Guillén F, Martínez MJ, Gutiérrez A, del Río JC (2005) Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol 8:195–204
Mei X, Dai Z, Fagherazzi S, Chen J (2016) Dramatic variations in emergent wetland area in China's largest freshwater lake, Poyang Lake. Adv Water Resour 96:1–10
Mishra S, Lee WA, Hooijer A, Reuben S, Sudiana IM, Idris A, Swarup S (2014) Microbial and metabolic profiling reveal strong influence of water table and land-use patterns on classification of degraded tropical peatlands. Biogeosciences 11:14009–14042
Moran JJ, House CH, Vrentas JM, Freeman KH (2008) Methyl sulfide production by a novel carbon monoxide metabolism in Methanosarcina acetivorans. Appl Environ Microbiol 74:540–542
Needham DM, Fuhrman JA (2016) Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 1(4):16005
Nunes FL, Aquilina L, De RJ, Francez AJ, Quaiser A, Caudal JP, Vandenkoornhuyse P, Dufresne A (2015) Time-scales of hydrological forcing on the geochemistry and bacterial community structure of temperate peat soils. Sci Rep 5:14612
Razavi BS, Liu S, Kuzyakov Y (2017) Hot experience for cold-adapted microorganisms: temperature sensitivity of soil enzymes. Soil Biol Biochem 105:236–243
Ren C, Wang T, Xu Y, Deng J, Zhao F, Yang G, Han X, Feng Y, Ren G (2018) Differential soil microbial community responses to the linkage of soil organic carbon fractions with respiration across land-use changes. For Ecol Manag 409:170–178
Rheindt FE, Székely T, Edwards SV, Lee PLM, Burke T, Kennerley PR, Bakewell DN, Alrashidi M, Kosztolányi A, Weston MA (2011) Conflict between genetic and phenotypic differentiation: the evolutionary history of a ‘lost and rediscovered’ shorebird. PLoS One 6:e26995
Romano N, Santini A (2002) Methods of soil analysis: part 4 physical methods. Journal Awwa
Rossi PG, Laurion I, Lovejoy C (2013) Distribution and identity of bacteria in subarctic permafrost thaw ponds. Aquatic Microbial Ecol 69:231–245
Saiyacork KR, Sinsabaugh RL, Zak DR (2002) The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol Biochem 34:1309–1315
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Shao X, Yang W, Wu M (2015) Seasonal dynamics of soil labile organic carbon and enzyme activities in relation to vegetation types in Hangzhou Bay Tidal Flat Wetland. PLoS One 10:e0142677
Shepard CC, Crain CM, Beck MW (2011) The protective role of coastal marshes: a systematic review and meta-analysis. PLoS One 6:e27374–e27374
Sinsabaugh RL, Saiyacork K, Long T (2003) Soil microbial activity in a Liquidambar plantation unresponsive to CO2-driven increases in primary production. Appl Soil Ecol 24(3):263–271
Spring S, Schulze R, Overmann J, Schleifer KH (2000) Identification and characterization of ecologically significant prokaryotes in the sediment of freshwater lakes: molecular and cultivation studies. FEMS Microbiol Rev 24:573–590
Stark CH, Condron LM, O’Callaghan M, Stewart A, Di HJ (2008) Differences in soil enzyme activities, microbial community structure and short-term nitrogen mineralisation resulting from farm management history and organic matter amendments. Soil Biol Biochem 40:1352–1363
Tiner RW (2010) NWIPlus: geospatial database for watershed-level functional assessment. Natl Wetl Newsl 32:4–7
Trivedi P, Delgadobaquerizo M, Trivedi C, Hu H, Anderson IC, Jeffries TC, Zhou J, Singh BK (2016) Microbial regulation of the soil carbon cycle: evidence from gene–enzyme relationships. ISME J 10:2593–2604
Truu M, Juhanson J, Truu J (2009) Microbial biomass, activity and community composition in constructed wetlands. Sci Total Environ 407:3958–3971
Wallenius K, Rita H, Mikkonen A, Lappi K, Lindström K, Hartikainen H, Raateland A, Niemi RM (2011) Effects of land use on the level, variation and spatial structure of soil enzyme activities and bacterial communities. Soil Biol Biochem 43:1464–1473
Wang L, Coles N, Wu C, Wu J (2014a) Effect of long-term reclamation on soil properties on a coastal plain, Southeast China. J Coast Res 30:661–669
Wang W, Liu H, Li Y, Su J (2014b) Development and management of land reclamation in China. Ocean Coast Manag 102:415–425
Wang J, Wang H, Cao Y, Bai Z, Qin Q (2016a) Effects of soil and topographic factors on vegetation restoration in opencast coal mine dumps located in a loess area. Sci Rep 6:22058
Wang NF, Zhang T, Yang X, Wang S, Yu Y, Dong LL, Guo YD, Ma YX, Zang JY (2016b) Diversity and composition of bacterial community in soils and lake sediments from an arctic lake area. Front Microbiol 7:1170
Ward NL, Challacombe JF, Janssen PH, Bernard H, Coutinho PM, Martin W, Gary X, Haft DH, Michelle S, Jonathan B (2009) Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75:2046–2056
Weise L, Ulrich A, Moreano M, Gessler A, Kayler ZE, Steger K, Zeller B, Rudolph K, Knezevicjaric J, Premke K (2016) Water level changes affect carbon turnover and microbial community composition in lake sediments. FEMS Microbiol Ecol 92:fiw035
Xiao W, Chen X, Jing X, Zhu B (2018a) A meta-analysis of soil extracellular enzyme activities in response to global change. Soil Biol Biochem 123:21–32
Xiao Y, Kong F, Yun X, Zhou W, Wang J, Hua Y, Zhang G, Zhao J (2018b) Comparative biogeography of the gut microbiome between Jinhua and landrace pigs. Sci Rep 8:5985
Xu S, Wang Y, Guo C, Zhang Z, Shang Y, Chen Q, Wang ZL (2016) Comparison of microbial community composition and diversity in native coastal wetlands and wetlands that have undergone long-term agricultural reclamation. Wetlands:1–10
Yakovchenko V, Sikora LJ, Kaufman DD (1996) A biologically based indicator of soil quality. Biol Fertil Soils 21:245–251
Yang S, Liebner S, Alawi M, Ebenhöh O, Wagner D (2014) Taxonomic database and cut-off value for processing mcrA gene 454 pyrosequencing data by MOTHUR. J Microbiol Method 103:3–5
Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR (2017) Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:14213
Acknowledgments
We thank Jeremy Kamen, MSc, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This research was supported by the National Natural Science Foundations of China (Grant No. 31660149, 31360127 and 30860062) and Nanchang University Seed Grant 358 for Biomedicine (9202-0210210807).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Yuan Ge
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
The rarefaction curves for natural wetland and four long-term reclaimed lands. Note: AW: abandoned wetland; VF: vegetable field; PS: pond sediment; RF: rice paddy field; NW: natural wetland (PNG 29 kb)
Fig. S2
A 100% stacked column chart of relative abundances of the dominant bacterial phyla derived from 16S rRNA genes at five sampling sites. The relative abundance of the targeted sequences to the total high-quality bacterial sequences (abundance > 1%) from natural wetland and four long-term reclaimed lands. Note: AW: abandoned wetland; VF: vegetable field; PS: pond sediment; RF: rice paddy field; NW: natural wetland (PNG 25 kb)
Fig. S3
The PCA analysis of soil extracellular enzyme from natural wetland and four long-term reclaimed lands. Note: AW: abandoned wetland; VF: vegetable field; PS: pond sediment; RF: rice paddy field; NW: natural wetland. Phox: phenol oxidase; Pero: peroxidase; Bglu: β-D-glucosidase; Bxyl: β-D-xylanase; NAG: N-acetyl-β-D-glucosaminidase; Phos: phosphatase (PNG 36 kb)
Rights and permissions
About this article
Cite this article
Huang, L., Hu, W., Tao, J. et al. Soil bacterial community structure and extracellular enzyme activities under different land use types in a long-term reclaimed wetland. J Soils Sediments 19, 2543–2557 (2019). https://doi.org/10.1007/s11368-019-02262-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-019-02262-1