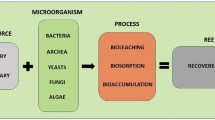
Abstract
Arsenic (As) is often found naturally as the co-contaminant in the uranium (U)-contaminated area, obstructing the bioremediation process. Although the U-contaminated environment harbors microorganisms capable of interacting with U which could be exploited in bioremediation. However, they might be unable to perform with their full potential due to As toxicity. Therefore, potential in arsenic resistance and oxidation is greatly desired among the microorganisms for a continued bioremediation process. In this study, arsenic-resistant bacteria were isolated from U ore collected from Bundugurang U mine, characterized and their As oxidation and U removal potentials were determined. 16S rRNA gene sequencing and phylogenetic analysis showed the affiliation of isolated bacteria with Microbacterium, Micrococcus, Shinella, and Bacillus. Except Bacillus sp. EIKU7, Microbacterium sp. EIKU5, Shinella sp. EIKU6, and Micrococcus sp. EIKU8 were found to resist more than 400 mM As(V); however, all the isolates could survive in 8 mM As(III). The isolates were found to readily oxidize As under different culture conditions and are also resistant towards Cd, Cr, Co, Ni, and Zn. All the isolates could remove more than 350 mg U/g dry cells within 48 h which were found to be highly dependent upon the concentration of U, biomass added initially, and on the time of exposure. Ability of the isolates to grow in nitrogen-free medium indicated that they can flourish in the nutrition deprived environment. Therefore, the recovery of isolates with the potent ability to resist and oxidize As from U ore might play an important role in toxic metal bioremediation including U.




Similar content being viewed by others
References
Abou-Shanab RA, vanBerkum P, Angle JS (2007) Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere 68:360–367
Acharya C (2015) Uranium bioremediation: approaches and challenges. In: Sukla L, Pradhan N, Panda S, Mishra B (eds) environmental microbial biotechnology. Soil Biol 45, Springer, Cham. https://doi.org/10.1007/978-3-319-19018-1_7
Ahn J, Park Y, Kim J-Y, Kim K-W (2005) Mineralogical and geochemical characterization of arsenic in an abandoned mine tailings of Korea. Environ Geochem Health 27:147–157
Bachate SP, Khapare RM, Kodam KM (2012) Oxidation of arsenite by two β-proteobacteria isolated from soil. Appl Microbiol Biotechnol 93:2135–2145
Bissen M, Frimmel FH (2003) Arsenic-a review. Part I: occurrence, toxicity, speciation, and mobility. Acta Hydrochim Hydrobiol 31:9–18
Bondici VF, Lawrence JR, Khan NH, Hill JE, Yergeau E, Wolfaardt GM, Warner J, Korber DR (2013) Microbial communities in low permeability, high pH uranium mine tailings: characterization and potential effects. J Appl Microbiol 114:1671–1686
Cheng KL, Hogan A, Parry DL, Markich S, Harford AJ, van Dam R (2010) Uranium toxicity and speciation during chronic exposure to the tropical freshwater fish, Mogurnda mogurnda. Chemosphere 79:547–554
Cheng Y, Holman HY, Lin Z (2012) Remediation of chromium and uranium contamination by microbial activity. Elements 8:107–112
Choudhary S, Sar P (2011) Identification and characterization of uranium accumulation potential of a uranium mine isolated Pseudomonas strain. World J Microbiol Biotechnol 27:1795–1801
Choudhary S, Sar P (2015) Interaction of uranium (VI) with bacteria: potential applications in bioremediation of U contaminated oxic environments. Rev Environ Sci Biotechnol 14:347–355
Choudhary S, Sar P (2016) Real-time PCR based analysis of metal resistance genes in metal resistant Pseudomonas aeruginosa strain J007. J Basic Microbiol. https://doi.org/10.1002/jobm.201500364
Corkhill C, Crean D, Bailey D, Makepeace C, Stennett M, Tappero R, Grolimund D, Hyatt N (2017) Multi-scale investigation of uranium attenuation by arsenic at an abandoned uranium mine, South Terras. npj Mater Degrad 1:19. https://doi.org/10.1038/s41529-017-0019-9
Cumberland SA, Douglas G, Grice K, Moreau JW (2016) Uranium mobility in organic matter-rich sediments: a review of geological and geochemical processes. Earth-Sci Rev 159:160–185
Drewniak L, Styczek A, Lopatka MM, Sklodowska A (2008) Bacteria, hypertolerant to arsenic in the rocks of an ancient gold mine, and their potyential role in dissemination of arsenic pollution. Environ Pollut 156:1069–1074
Fey DL, Desborough GA (2004) Metal leaching in mine-waste materials and two schemes for classification of potential environmental effects of mine-waste piles. In: D.A. Nimick, S.E. Church and S.E. Finger (Editors), Integrated investigations of environmental effects of historical Mining in the Basin and Boulder Mining Districts, Boulder River watershed, Jefferson County, Montana. U.S. Geological Survey, Denver, pp. 530
Gadd GM (2010) Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 156:609–643
Garbeva P, van Veen J, van Elsas J (2003) Predominant Bacillus spp. in agricultural soil under different management regimes detected via PCR-DGGE. Microb Ecol 45:302. https://doi.org/10.1007/s00248-002-2034-8
Gerber U, Hübner R, Rossberg A, Krawczyk-Bärsch E, Merroun ML (2018) Metabolism-dependent bioaccumulation of uranium by Rhodosporidium toruloidesisolated from the flooding water of a former uranium mine. PLoS One 13:e0201903. https://doi.org/10.1371/journal.pone.020190
Goswami D, Parmar S, Vaghela H, Dhandhukia P, Thakker J (2015) Describing Paenibacillus mucilaginosus strain N3 as an efficient plant growth promoting rhizobacteria (PGPR). Cogent Food Agric 1:1000714. https://doi.org/10.1080/23311932.2014.1000714
Islam E, Sar P (2011) Culture-dependent and -independent molecular analysis of bacterial community within uranium ore. J Basic Microbiol 4:1–13
Islam E, Sar P (2016) Diversity, metal resistance and uranium sequestration abilities of bacteria from uranium ore deposit in deep earth stratum. Ecotoxicol Environ Saf 127:12–21
Islam E, Dhal PK, Kazy SK, Sar P (2011) Molecular analysis of bacterial communities in uranium ores and surrounding soils from Banduhurang open cast uranium mine, India: a comparative study. J Environ Sci Health A 46:271–280
Islam E, Paul D, Sar P (2014) Microbial diversity in uranium deposits from Jaduguida and Bagjata mines, India as revealed bt clone library and denaturing gradient gel electrophoresis analyses. Geomicrobiol J 31:862–874
Jackson CR, Dugas SL (2003) Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol Biol 3:3–18
Jakubick A, Gatzweile R, Mager D, Mac A, Robertson G (1997) The Wismut waste rock pile remediation programme of the Ronneburg district, Germany, Abstr. 4th International Conference on Acid Rock Drainage, Vancouver B.C. 1285-1301
Kazy SK, D’Souza SF, Sar P (2009) Uranium and thorium sequestration by a Pseudomonas sp.: mechanism and chemical characterization. J Hazard Mater 163:65–72
Kolhe N, Zinjarde S, Acharya C (2018) Responses exhibited by various microbial groups relevant to uranium exposure. Biotechnol Adv 36:1828–1846
Kostka JE, Green SJ (2011) Microorganisms and processes linked to uranium reduction and immobilization. In: Stolz J, Oremland R (eds) Microbial metal and metalloid metabolism. ASM Press, Washington, pp 117–138
Kulkarni S, Misra CS, Gupta A, Ballal A, Apte SK (2016) Interaction of uranium with bacterial cell surfaces: inferences from phosphatase-mediated uranium precipitation. Appl Environ Microbiol 82:4974–4983
Kumar R, Acharya V, Singh D, Kumar S (2018) Strategies for high-altitude adaptation revealed from high-quality draft genome of non-violacein producing Janthinobacterium lividum ERGS5:01. Stand Genomic Sci 13:11. https://doi.org/10.1186/s40793-018-0313-3
Lim KT, Shukor MY, Wasoh H (2014) Physical, chemical, and biological methods for the removal of arsenic compounds. Biomed Res Int 2014:9–9. https://doi.org/10.1155/2014/503784
Lloyd J (2003) Microbial reduction of metals and radionuclides. FEMS Microbiol Rev 27:411–425
Martinez RJ, Wang Y, Raimondo MA, Coombs JM, Barkay T, Sobecky PA (2006) Horizontal gene transfer of PIB-type ATPases among bacteria isolated from radionuclide and metal-contaminated subsurface soils. Appl Environ Microbiol 72:3111–3118
Meharg AA, Hartley-Whitaker J (2002) Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol 154:29–43
Moldovan BJ, Jiang DT, Hendry MJ (2003) Mineralogical characterization of arsenic in uranium mine tailings precipitated from iron-rich hydrometallurgical solutions. Environ Sci Technol 37:873–879
Morcillo F, Gonźalez-Muñoz MT, Reitz T, Romero-Gonźalez ME, Arias JM, Merroun ML (2014) Biosorption and biomineralization of U(VI) by the marine bacterium Idiomarina loihiensis MAH1: effect of background electrolyte and pH. PLoS One 9:e91305. https://doi.org/10.1371/journal.pone.0091305
Mouser PJ, N’Guessan AL, Elifantz H, Holmes DE, Williams KH, Wilkins MJ, Long PE, Lovley DR (2009) Influence of heterogeneous ammonium availability on bacterial community structure and the expression of nitrogen fixation and ammonium transporter genes during in situ bioremediation of uraniumcontaminated groundwater. Environ Sci Technol 43:4386–4392
Nedelkova M, Merroun ML, Rossberg A, Hennig C, Selenska-Pobell S (2007) Microbacterium isolates from the vicinity of a radioactive waste depository and their interactions with uranium. FEMS Microbiol Ecol 59:694–705
Newsome L, Morris K, Lloyd JR (2014) The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem Geol 363:164–184
Nilgiriwala KS, Alahari A, Rao AS, Apte SK (2008) Cloning and overexpression of alkaline phosphatase SPAP from Sphingomonas sp. strain BSAR-1 for bioprecipitation of uranium from alkaline solutions. Appl Environ Microbiol 74:5516–5523
Oremland RS, Stolz JF (2005) Arsenic, microbes and contaminated aquifers. Trends Microbiol 13:45–49
Paul D, Poddar S, Sar P (2014) Characterization of arsenite oxidizing bacteria isolated from arsenic-contaminated ground water of West Bengal. J Environ Sci Health A 49:1481–1492
Pichler T, Hendry MJ, Hall GEM (2001) The mineralogy of arsenic in uranium mine tailings at the rabbit lake in-pit facility, northern Saskatchewan, Canada. Environ Geol 40:495–506
Qiu J, Yang Y, Zhang J, Wang H, Ma Y, He J, Lu Z (2016) The complete genome sequence of the nicotine-degrading bacterium Shinella sp. HZN7. Front Microbiol 7:1348. https://doi.org/10.3389/fmicb.2016.01348
Schiewer S, Volesky B (2000) Biosorption processes for heavy metal removal. In: Lovley DR (ed) Environmental microbe–metal interactions. ASM Press, Washington D. C, pp 329–362
Simeonova DD, Lievremont D, Lagarde F, Muller DA, Groudeva VI, Lett MC (2004) Microplate screening assay for the detection of arsenite-oxidizing and arsenate-reducing bacteria. FEMS Microbiol Lett 237:249–253
Suzuki Y, Banfield JF (2004) Resistance to, and accumulation of, uranium by bacteria from a uranium-contaminated site. Geomicrobiol J 21:113–121
Tabak HH, Lens PNL, van Hullebusch ED, Dejonghe W (2005) Developments in bioremediation of soils and sediments polluted with metals and radionuclides: 1. Microbial processes and mechanisms affecting bioremediation of metal contamination and influencing metal toxicity and transport. Rev Environ Sci Biotechnol 4:115–156
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Vazquez GJ, Dodge CJ, Francis AJ (2007) Interactions of uranium with polyphosphate. Chemosphere 70:263–269
Wall JD, Krumholz LR (2006) Uranium reduction. Annu Rev Microbiol 60:149–166
Whiteley AS, Bailey MJ (2000) Bacterial community structure and physiological state within an industrial phenol bioremediation system. Appl Environ Microbiol 66:2400–2407
Wolfaardt GM, Hendry MJ, Korber DR (2008) Microbial distribution and diversity in saturated high pH, uranium mine tailings, Saskatchewan, Canada. Can J Microbial 54:932–940
Yong P, Eccles H, Macaskie LE (1996) Determination of uranium, thorium and lanthanum in mixed solutions using simultaneous spectrophotometry. Anal Chim Acta 329:173–179
Acknowledgements
We thankfully acknowledge Kristen Powers and Dr. Sanjoy Kumar Khan, Developmental Biology, Harvard School of Dental Medicine, Boston, MA for language editing of the script.
Funding
This study was financially supported by DST-WTI grant (Grant No. DST/TM/WTI/2K16/276/(G)), Government of India. A support from DST-PURSE, University of Kalyani is also highly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Diane Purchase
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• Arsenic-resistant bacteria were isolated from U ore and characterized for U removal
• Isolated bacteria were affiliated to Microbacterium, Micrococcus, Shinella, and Bacillus
• All the bacteria were able to resist more than 400 mM As(V) and 8 mM As(III)
• Isolates removed more than 350 mg U/g dry cells and exhibit As (III) oxidation potential
• Heavy metal-resistant and nitrogen fixing ability indicated their activity in harsh condition
Electronic supplementary material
ESM 1
(DOC 49 kb)
Rights and permissions
About this article
Cite this article
Bhakat, K., Chakraborty, A. & Islam, E. Characterization of arsenic oxidation and uranium bioremediation potential of arsenic resistant bacteria isolated from uranium ore. Environ Sci Pollut Res 26, 12907–12919 (2019). https://doi.org/10.1007/s11356-019-04827-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-04827-6