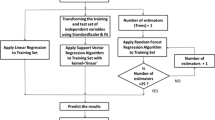
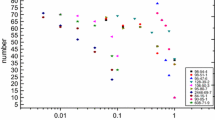
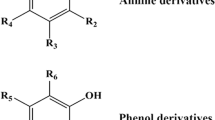
Abstract
In this work, a new norm descriptor is proposed based on atomic properties. A quantitative structure-activity relationship (QSAR) model for predicting the toxicity of organic compounds to fathead minnow is further developed by norm descriptors. Results indicate that this new model based on the norm descriptors has satisfactory predictive results with the squared correlation coefficient (R2) and squared relation coefficient of the cross validation (Q2) of 0.8174 and 0.7923, respectively. Combining with Y-randomization test, applicability domain test, and comparison with other references, calculation results indicate that the QSAR model performs well both in the stability and the accuracy with wide application domain, which might be further used effectively for the safe and risk assessment of various organics.






Similar content being viewed by others
References
Anonymous ADAPT
Anonymous CODESSA
Anonymous Dragon
Barron MG, Lilavois CR, Martin TM (2015) MOAtox: a comprehensive mode of action and acute aquatic toxicity database for predictive model development. Aquat Toxicol 161:102–107
Basant N, Gupta S (2017) QSAR modeling for predicting mutagenic toxicity of diverse chemicals for regulatory purposes. Environ Sci Pollut Res 24:14430–14444
Belanger SE, Brill JL, Rawlings JM, Price BB (2016) Development of acute toxicity quantitative structure activity relationships (QSAR) and their use in linear alkylbenzene sulfonate species sensitivity distributions. Chemosphere 155:18–27
Cassani S, Kovarich S, Papa E, Roy PP, van der Wal L, Gramatica P (2013) Daphnia and fish toxicity of (benzo)triazoles: validated QSAR models, and interspecies quantitative activity–activity modelling. J Hazard Mater 258–259:50–60
Cassotti M, Ballabio D, Todeschini R, Consonni V (2015) A similarity-based QSAR model for predicting acute toxicity towards the fathead minnow (Pimephales promelas). SAR QSAR Environ Res 26:217–243
Colombo A, Benfenati E, Karelson M, Maran U (2008) The proposal of architecture for chemical splitting to optimize QSAR models for aquatic toxicity. Chemosphere 72:772–780
Drgan V, Župerl Š, Vračko M, Como F, Novič M (2016) Robust modelling of acute toxicity towards fathead minnow (Pimephales promelas) using counter-propagation artificial neural networks and genetic algorithm. SAR QSAR Environ Res 27:501–519
Fangyou Y, Wensi HE, Qingzhu J, Qiang W, Shuqian X, Peisheng MA. (2018) Prediction of ionic liquids viscosity at variable temperatures and pressures. Chem Eng Sci
Gramatica P (2007) Principles of QSAR models validation: internal and external. QSAR Comb Sci 26:694–701
He WS, Yan FY, Jia QZ, Xia SQ, Wang Q (2018) QSAR models for describing the toxicological effects of ILs against Staphylococcus aureus based on norm indexes. Chemosphere 195:831–838
Jagiello K, Mostrag-Szlichtyng A, Gajewicz A, Kawai T, Imaizumi Y, Sakurai T, Yamamoto H, Tatarazako N, Mizukawa K, Aoki Y, Suzuki N, Watanabe H, Puzyn T (2015) Towards modelling of the environmental fate of pharmaceuticals using the QSPR-MM scheme. Environ Model Softw 72:147–154
Jia QZ, Cui X, Li L, Wang Q, Liu Y, Xia SQ, Ma PS (2015) Quantitative structure-activity relationship for high affinity 5-HT1A receptor ligands based on norm indexes. J Phys Chem B 119:15561–15567
Jin X, Jin M, Sheng L (2014) Three dimensional quantitative structure–toxicity relationship modeling and prediction of acute toxicity for organic contaminants to algae. Comput Biol Med 51:205–213
Levet A, Bordes C, Clément Y, Mignon P, Chermette H, Marote P, Cren-Olivé C, Lantéri P (2013) Quantitative structure–activity relationship to predict acute fish toxicity of organic solvents. Chemosphere 93:1094–1103
Lyakurwa F, Yang X, Li X, Qiao X, Chen J (2014a) Development and validation of theoretical linear solvation energy relationship models for toxicity prediction to fathead minnow (Pimephales promelas). Chemosphere 96:188–194
Lyakurwa FS, Yang X, Li X, Qiao X, Chen J (2014b) Development of in silico models for predicting LSER molecular parameters and for acute toxicity prediction to fathead minnow (Pimephales promelas). Chemosphere 108:17–25
Martin TM, Young DM (2001) Prediction of the acute toxicity (96-h LC50) of organic compounds to the fathead minnow (Pimephales promelas) using a group contribution method. Chem Res Toxicol 14:1378–1385
Papa E, Battaini F, Gramatica P (2005) Ranking of aquatic toxicity of esters modelled by QSAR. Chemosphere 58:559–570
Ren YY, Zhou LC, Yang L, Liu PY, Zhao BW, Liu HX (2016) Predicting the aquatic toxicity mode of action using logistic regression and linear discriminant analysis. SAR QSAR Environ Res 27:721–746
Roy K, Das RN (2012) QSTR with extended topochemical atom (ETA) indices. 15. Development of predictive models for toxicity of organic chemicals against fathead minnow using second-generation ETA indices. SAR QSAR Environ Res 23:125–140
Roy K, Das RN, Popelier PLA (2015a) Predictive QSAR modelling of algal toxicity of ionic liquids and its interspecies correlation with Daphnia toxicity. Environ Sci Pollut Res 22:6634–6641
Roy K, Kar S, Das R (2015b): Chapter 7—validation of QSAR models. Understanding the basics of QSAR for applications in pharmaceutical sciences and risk assessment, Academic Press, 231–289
Rucker C, Rucker G, Meringer M (2007) y-Randomization and its variants in QSPR/QSAR. J Chem Inf Model 47:2345–2357
Russom CL, Bradbury SP, Broderius SJ, Hammermeister DE, Drummond RA (1997) Predicting modes of toxic action from chemical structure: acute toxicity in the fathead minnow (Pimephales promelas). Environ Toxicol Chem 16:948–967
Sabljić A, Piver WT (1992) Quantitative modeling of environmental fate and impact of commercial chemicals. Environ Toxicol Chem 11:961–972
Schuurmann G, Ebert RU, Kuhne R (2011) Quantitative read-across for predicting the acute fish toxicity of organic compounds. Environ Sci Technol 45:4616–4622
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb Sci 22:69–77
Verhaar HJM, Leeuwen CJV, Hermens JLM (1992) Classifying environmental pollutants. Chemosphere 25:471–491
Wang Q, Jia QZ, Yan LH, Xia SQ, Ma PS (2014) Quantitative structure-toxicity relationship of the aquatic toxicity for various narcotic pollutants using the norm indexes. Chemosphere 108:383–387
Wang Y, Yan F, Jia Q, Dai Y, Wang Q (2017): Quantitative structure-activity relationship of anti-HIV integrase and reverse transcriptase inhibitors using norm indexes. SAR QSAR Environ Res, 1–20
Wang YL, Yan FY, Jia QZ, Wang Q (2018) Quantitative structure-property relationship for critical micelles concentration of sugar-based surfactants using norm indexes. J Mol Liq 253:205–210
Wu X, Zhang Q, Hu J (2016) QSAR study of the acute toxicity to fathead minnow based on a large dataset. SAR QSAR Environ Res 27:147–164
Xu XY, Li L, Yan FY, Jia QZ, Wang Q, Ma PS (2016) Predicting solubility of fullerene C-60 in diverse organic solvents using norm indexes. J Mol Liq 223:603–610
Yali W, Fangyou Y, Qingzhu J, Qiang W (2017) Assessment for multi-endpoint values of carbon nanotubes: quantitative nanostructure-property relationship modeling with norm indexes. J Mol Liq:248
Yan FS, He WS, Jia QZ, Wang Q, Xia SQ, Ma PS (2018) Prediction of ionic liquids viscosity at variable temperatures and pressures. Chem Eng Sci 184:134–140
Yin JC, Jia QZ, Yan FY, Wang Q (2017) Predicting heat capacity of gas for diverse organic compounds at different temperatures. Fluid Phase Equilib 446:1–8
Yuan H, Wang Y-Y, Cheng Y-Y (2007) Mode of action-based local QSAR modeling for the prediction of acute toxicity in the fathead minnow. J Mol Graph Model 26:327–335
Funding
This work was supported by the National Natural Science Foundation of China (21676203 and 21808167).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Marcus Schulz
Electronic supplementary material
Table S1
(XLSX 74 kb)
Appendix
Appendix
The 364th molecule (acetic acid) was shown as an example and the molecular structure was as follows and structure of chemicals were optimized and obtained the most stable molecule structure.

Firstly, by Eqs. (1)–(4), the position matrices (Mm, m = 1,2,3,4) were calculated and shown as follow:
Then, by Eqs. (5)–(9), the property matrices (MEn, n = 1, 2, 3, 4, 5) were calculated and shown as follows:
-
ME1 = [5.6549 5.6549 4.3982 4.3982 3.7699 3.7699 3.7699 3.7699]T
-
ME2 = [2.5500 2.5500 3.4400 3.4400 2.2000 2.2000 2.2000 2.2000]T
-
ME3 = [0.0183 0.0067 0.1353 0.0498 0.1353 0.1353 0.1353 0.1353]T
-
ME4 = [0.2078 0.2078 0.2018 0.2018 0.2323 0.2323 0.2323 0.2323]T
-
ME5 = [1.0865 1.0865 1.3139 1.3139 1.3120 1.3120 1.3120 1.3120]T
The combinatorial matrices were performed by Eqs. (10)–(12). Finally, the value of the descriptors were obtained by Eqs. (13)–(15) and the largest eigenvalue of the matrix (MB,m,n) also need to calculate. The descriptors were shown in the Table 4 which were selected to predict the toxicity to fathead minnow.
Based on above, the pLC50 was calculated by Eq. (16).
The predicted toxicity value for fathead minnow was 2.690 mol/L, and the experimental value was 2.850 mol/L.
Rights and permissions
About this article
Cite this article
Jia, Q., Zhao, Y., Yan, F. et al. QSAR model for predicting the toxicity of organic compounds to fathead minnow. Environ Sci Pollut Res 25, 35420–35428 (2018). https://doi.org/10.1007/s11356-018-3434-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-018-3434-8