Abstract
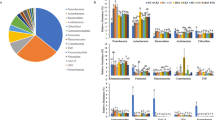
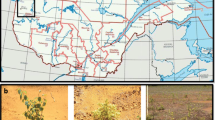
The planetary importance of microbial function requires urgently that our knowledge and our exploitation ability is extended, therefore every occasion of bioprospecting is welcome. In this work, bioprospecting is presented from the perspective of the UMBRELLA project, whose main goal was to develop an integral approach for remediation of soil influenced by mining activity, by using microorganisms in association with plants. Accordingly, this work relies on the cultivable fraction of microbial biodiversity, native to six mining sites across Europe, different for geographical, climatic and geochemical characteristics but similar for suffering from chronic stress. The comparative analysis of the soil functional diversity, resulting from the metabolic profiling at community level (BIOLOG ECOPlates) and confirmed by the multivariate analysis, separates the six soils in two clusters, identifying soils characterised by low functional diversity and low metabolic activity. The microbial biodiversity falls into four major bacterial phyla: Actinobacteria, Proteobacteria, Firmicutes and Bacteroidetes, including a total of 47 genera and 99 species. In each soil, despite harsh conditions, metabolic capacity of nitrogen fixation and plant growth promotion were quite widespread, and most of the strains showed multiple resistances to heavy metals. At species-level, Shannon’s index (alpha diversity) and Sørensen's Similarity (beta diversity) indicates the sites are indeed diverse. Multivariate analysis of soil chemical factors and biodiversity identifies for each soil well-discriminating chemical factors and species, supporting the assumption that cultured biodiversity from the six mining sites presents, at phylum level, a convergence correlated to soil factors rather than to geographical factors while, at species level, reflects a remarkable local characterisation.




Similar content being viewed by others
References
Amoroso MJ, Schubert D, Mitscherlich P, Schumann P, Kothe E (2000) Evidence for high affinity nickel transporter genes in heavy metal resistant Streptomyces species. J Basic Microbiol 40:295–301
Battistuzzi FU, Hedges SB (2009) A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol 26(2):335–343
Dworkin M, Foster J (1958) Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol 75:592–601
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. PNAS 103(3):626–631
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88(6):1354–1364
Forney LJ, Zhou X, Brown CJ (2004) Molecular microbial ecology: land of the one-eyed king. Curr Opin Microbiol 7:210–220
Garland JL (1996) Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem 28(2):213–221
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57(8):2351–2359
Gordon SA, Weber RP (1951) Colorimetric estimation of indoleacetic acid. Plant Physiol 26(1):192–195
Gupta R, Singal R, Shankar A, Kuhad RC, Saxena RK (1994) A modified plate assay for screening phosphate solubilizing microorganisms. J Gen Appl Microbiol 40:255–260
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes Appl. Environ Microbiol 72(3):1719–1728
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics: John Innes Foundation, Norwich Research Park, Colney, Norwich NR4 7UH, UK
Laplante K, Derome N (2011) Parallel changes in the taxonomical structure of bacterial communities exposed to a similar environmental disturbance. Ecol Evol 1(4):489–501
MacArthur R, Wilson E (1967) The theory of island biogeography. Princeton University Press, Princeton, New Jersey, USA
Milagres AM, Machuca A, Napoleão D (1999) Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J Microbiol Methods 37(1):1–6
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
O’Malley MA (2008) ‘Everything is everywhere: but the environment selects’: ubiquitous distribution and ecological determinism in microbial biogeography. Stud Hist Philos Biol Biomed Sci 39(3):314–325
Patten CL, Glick BR (2002) Role of Pseudomonas putida indole acetic acid in development of host plant root system. Appl Environ Microbiol 48:3795–3801
Pikovskaya RI (1948) Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Mikrobiologia 17:362–370
Rappè MS, Giovannoni SJ (2003) The uncultured microbial majority. Annu Rev Microbiol 57:369–394
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schmidt T, Schlegel HG (1989) Nickel and cobalt resistance of various bacteria isolated from soil and highly polluted domestic and industrial wastes. FEMS Microb Ecol 62:315–328
Sprocati AR, Alisi C, Tasso F, Marconi P, Sciullo A, Pinto V, Chiavarini S, Ubaldi C, Cremisini C (2012) Feasibility study for bioremediation of a soil co-contaminated by diesel oil and heavy metals using a tailor-made microbial formula as bioaugmentation agent. Process Biochem 47:1649–1655
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Yao H, He Z, Wilson MJ, Campbell CD (2000) Microbial biomass and community structure in a sequence of soils with increasing fertility and changing land use. Microb Ecol 40:223–237
Whittaker RH (1972) Evolution and measurement of species diversity. Taxonomy 21:213–251
Zinder SH, Salyers AA (2001) Microbial ecology—new directions, new importance. In: Boone DR, Castenholz RW (eds) Bergey's manual of systematic bacteriology vol. 1: the Archaea and the deeply branching and phototrophic bacteria. Springer-Verlag, New York, pp 101–109
Acknowledgements
This study was supported by the EU 7FP-UMBRELLA (EU 226870).
The authors warmly thank Federico Marini from the University Sapienza (Rome, Italy) for the help in statistical analysis
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Robert Duran
Rights and permissions
About this article
Cite this article
Sprocati, A.R., Alisi, C., Tasso, F. et al. Bioprospecting at former mining sites across Europe: microbial and functional diversity in soils. Environ Sci Pollut Res 21, 6824–6835 (2014). https://doi.org/10.1007/s11356-013-1907-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-013-1907-3