Abstract
Background aim and scope
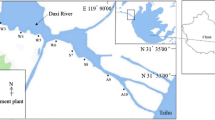
Though the tidal Anacostia River, a highly polluted riverine system, has been well characterized with regard to contaminants, its overall resident bacterial populations have remained largely unknown. Improving the health of this system will rely upon enhanced understanding of the diversity and functions of these communities. Bacterial DNA was extracted from archived (AR, year 2000) and fresh sediments (RE, year 2006) collected from various locations within the Anacostia River. Using a combination of metabolic and molecular techniques, community snapshots of sediment bacterial diversity and activity were produced.
Results
Employing Biolog EcoPlates, metabolic analysis of RE sediments from July revealed similar utilization of amines, amino acids, carbohydrates, carboxylic acids, and polymers at all sites. Normalized optical density measurements demonstrated that for most compounds, utilizations were similar though when differences did occur, the downstream site was enhanced compared to one or both of the upstream sites. Using denaturing gradient gel electrophoresis, bacterial diversity fingerprints of operational taxonomic units (OTUs) were obtained. Dendograms of the banding patterns revealed qualitative relationships as well as differences between replicate samples from similar sites. Replicates from the AR sites shared several common OTUs, while RE sites were more varied. Species richness and Shannon diversity indices generally increased with increasingly downstream locations, and were significant for the AR sediments (analysis of variance, P < 0.0001). Carbon and nitrogen content and concentration of fine grain sediment (<63 μm) were positively correlated with OTU richness (r 2 = 0.37, P = 0.0008; r 2 = 0.45, P < 0.0001; r 2 = 0.48, P = 0.001, respectively).
Conclusions
This study demonstrated that the bacterial communities from all regions sampled were not only metabolically active with the capacity to utilize several different compounds as energy sources but also were genetically diverse. This study is the first to focus on the overall bacterial community, providing insight into this vital component of stream ecosystems. Understanding the bacterial components of aquatic systems such as the Anacostia River will increase our knowledge of the overall structure and function of the ecological communities in polluted systems, subsequently enhancing our ability to improve the health of this important tidal river.




Similar content being viewed by others
References
Bai S, Lung W-S (2006) Three-dimensional modeling of fecal coliform in the tidal basin and Washington Channel, Washington, DC. J Environ Sci Health Part A 41:1327–1346
Christian BW, Lind OT (2006) Key issues concerning Biolog use for aerobic and anaerobic freshwater bacterial community-level physiological profiling. Int Rev Hydrobiol 91:257–268
Cody DG, Heath RT, Leff LG (2000) Characterization of benthic bacterial assemblages in a polluted stream using denaturing gradient gel electrophoresis. Hydrobiol 432:207–215
Ewers EC (2009) Polycyclic aromatic hydrocarbon-degrading bacteria in Anacostia River sediments: presence and metabolic capabilities. Master’s Thesis
Ferris MJ, Muyzer G, Ward DM (1996) Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62:340–346
Findlay SEG, Sinsabaugh RL (1999) Unraveling the sources and bioavailability of dissolved organic matter in aquatic ecosystems. Mar Freshw Res 50:781–790
Fortunato CS, Carlini DB, Ewers E, Bushaw-Newton KL (2009) Nitrifier and denitrifier molecular operational taxonomic unit compositions from a freshwater estuary of Chesapeake Bay. Can J Microbiol 55:333–346
Foster GD, Roberts EC Jr, Gruessner B, Velinsky DJ (2000) Hydrogeochemistry and transport of organic contaminants in an urban watershed of Chesapeake Bay (USA). Appl Geochem 15:901–915
Fredrickson JK, Balkwill DL (1998) Sampling and environmental techniques. In: Burlage RS et al (eds) Techniques in microbial ecology. Oxford University Press, Oxford, pp 239–254
Garland JL, Lehman RM (1999) Dilution/extinction of community phenotypic characters to estimate relative structural diversity in mixed communities. FEMS Microbiol Ecol 30:333–343
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359
Hwang HM, Foster GD (2008) Polychlorinated biphenyls in stormwater runoff entering the tidal Anacostia River, Washington, DC, through small urban catchments and combined sewer outfalls. J Environ Sci Health A 43:567–576
Insam H (1997) A new set of substrates proposed for community characterization of environmental samples. In: Insam H, Rangger A (eds) Microbial communities, functional versus structural approaches. Springer, Berlin, pp 260–261
Kjellerup BV, Sun X, Ghosh U, May HD, Sowers KR (2008) Site-specific microbial communities in three PCB-impacted sediments are associated with different in situ dechlorinating activities. Environ Microbiol 10:1296–1309
Labbé D, Margesin R, Schinner F, Whyte LG, Greer CW (2007) Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated Alpine soils. FEMS Microbiol Ecol 59:466–475
Li D, Yang M, Li Z, Qi R, He J, Liu H (2008) Change of bacterial communities in sediments along Songhua River in Northeastern China after a nitrobenzene pollution event. FEMS Microbiol Ecol 65:494–503
MacAvoy SE, Ewers EC, Bushaw-Newton KL (2009) Nutrients, oxygen dynamics, stable isotopes and fatty acid concentrations of a freshwater tidal system, Washington, D.C. J Environ Monit 11:1622–1628
McGee BL, Pinkney AE, Velinsky DJ, Ashley JTF, Fisher DJ, Ferrington LC, Norberg-King TJ (2009) Using the sediment quality triad to characterize baseline conditions in the Anacostia River, Washington, DC, USA. Environ Monit Assess 156:51–67
Metropolitan Washington Council of Governments (MWCOG) (1998) Anacostia watershed restoration progress and conditions report 1990–1997. MWCOG, Washington, DC
Münster U, De Haan H (1998) The role of microbial extracellular enzymes. In: Hessen DO, Tranvik LJ (eds) Aquatic humic substances: ecology and biogeochemistry. Springer, Berlin, pp 199–257
Muyzer G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2:317–322
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Phelps HL (1985) Summer 1984 survey of mollusk populations of the Potomac and Anacostia Rivers near Washington DC. Report to the District of Columbia Environmental Services, Washington, DC
Phelps HL (1993) Sediment toxicity of the Anacostia River estuary, Washington DC. Bull Environ Contam Toxicol 51:582–587
Phelps HL (2004) Sources of bioavailable toxic pollutants in the Anacostia watershed. Part III, DC Water Resources Research Center Report 2003DC34B. DC Water Resources Research Institute, Washington, DC
Pinkney AE, Harshbarger JC, May EB, Reichert WL (2004) Tumor prevalence and biomarkers of exposure and response in brown bullhead (Ameiurus nebulosus) from the Anacostia River, Washington, DC and Tuckahoe River, Maryland. Environ Toxicol Chem 24:638–647
Rattner BA, Golden NH, Toschik PC, McGowan PC, Custer TW (2008) Concentrations of metals in blood and feathers of nesting ospreys (Pandion haliaetus) in Chesapeake and Delaware Bays. Arch Environ Contam Toxicol 54:114–122
Ros M, Pascual JA, Moreno JL, Hernandez MT, Garcia C (2009) Evaluation of microbial community activity, abundance, and structure in a semiarid soil under cadmium pollution at laboratory level. Water Air Soil Pollut 203:229–242
Sala MM, Terrado R, Lovejoy C, Unrein F, Pedros-Alio C (2008) Metabolic diversity of heterotrophic bacterioplankton over winter and spring in the coastal Arctic Ocean. Environ Microbiol 10:942–949
Shala L, Foster GD (2010) Surface water concentrations and loading budgets of pharmaceuticals and other domestic-use chemicals in an urban watershed (Washington, DC, USA). Arch Env Contam Toxicol 58:551–561
Sinsabaugh RL, Foreman CM (2001) Activity profiles of bacterioplankton in a eutrophic river. Freshw Biol 46:1239–1249
Sinsabaugh RL, Findlay S, Franchini P, Fischer D (1997) Enzymatic analysis of riverine bacterioplankton production. Limnol Oceanogr 42:29–38
Smalla K, Wachtendorf U, Holger H, Liu WT, Forney L (1998) Analysis of Biolog GN substrate utilization patterns by microbial communities. Appl Environ Microbiol 64:1220–1225
Syracuse Research Corporation (SRC) (2000) Interpretive summary of existing data relevant to potential contaminants of concern within the Anacostia River watershed. SRC, New York
Velinsky DJ, Ashley JTF (2001) Deposition and spatial distribution of sediment-bound contaminants in the Anacostia River, District of Columbia, Report No. 01-30D. Academy of Natural Sciences, Philadelphia
Velinsky DJ, Wade TL, Schlekat CE, McGee BL, Presley BJ (1994) Tidal river sediments in the Washington, D.C. area. I. Distribution and sources of trace metals. Estuaries 17:305–320
Velinsky DJ, Riedel GF, Ashley JTF, Cornwell JC (2011) Historical contamination of the Anacostia River, Washington, D.C. Environ Monit Assess 183:307–328
Wade TL, Velinsky DJ, Reinharz E, Schlekat CE (1994) Tidal river sediments in the Washington DC area II. Distribution and sources of organic contaminants. Estuaries 17:321–333
Yu Z, Morrison M (2004) Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl Enivron Microbiol 70:4800–4806
Zar JH (1999) Biostatistical analysis, 4th edn. Prentice-Hall, Upper Saddle River
Acknowledgments
We thank Emily Adams, Caroline Fortunato, and Ken Jensen for excellent field and laboratory assistance. Andrew Newton, as always, provided great support. Two anonymous reviewers provided excellent comments on an earlier draft. This research was supported by grants from the US Geological Survey Water Resources Research Institute and American University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Bushaw-Newton, K.L., Ewers, E.C., Velinsky, D.J. et al. Bacterial community profiles from sediments of the Anacostia River using metabolic and molecular analyses. Environ Sci Pollut Res 19, 1271–1279 (2012). https://doi.org/10.1007/s11356-011-0656-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-011-0656-4