Abstract
Introduction
Members of the genus Sphingomonas have raised increasing attention due to their ability for polycyclic aromatic hydrocarbon (PAH) degradation and their ubiquity in the environment. However, few studies have revealed the ecological information on the abundance and diversity of Sphingomonas in the environment.
Materials and methods
A primer set targeting the Sphingomonas 16S rRNA gene was designed. The specificity was tested with four petroleum-contaminated soils by construction of clone libraries and further restriction fragment length polymorphism analysis. Subsequently, real time PCR and denaturing gradient gel electrophoresis (DGGE) assays were used to evaluate the abundance and diversity of Sphingomonas in the Shenfu irrigation zone, China.
Results
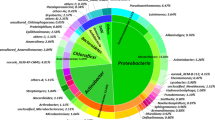
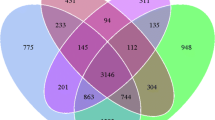
A genus-specific primer set SA/429f-933r was developed, and 90% of the sequences retrieved from soil clone libraries were related to Sphingomonas. Members of the genus Sphingomonas were detected in all soils, and significant correlation (p < 0.05) was observed between the Sphingomonas abundance and the ratios of PAHs to total petroleum hydrocarbon (TPH). DGGE profiles revealed Sphingomonas population structures differed greatly in different sites. The Sphingomonas diversity was not statistically (p > 0.05) correlated with the contamination level. Some of the soil-derived sequences were not grouped phylogenetically with sequences of known Sphingomonas, indicating new members of the Sphingomonas genus might be present in the Shenfu irrigation zone.
Conclusion
The newly designed Sphingomonas-selective primers were specific and practicable for analyzing Sphingomonas abundance and diversity in petroleum-contaminated soils. The significant correlation between the abundance and the ratios of PAHs to TPH suggested an important role of Sphingomonas in PAH bioremediation.




Similar content being viewed by others
References
Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bastiaens L, Springael D, Wattiau P, Harms H, DeWachter R, Verachtert H et al (2000) Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl Environ Microbiol 66:1834–1843
Brito E, Guyoneaud R, Go i-Urriza M, Ranchou-Peyruse A, Verbaere A, Crapez M et al (2006) Characterization of hydrocarbonoclastic bacterial communities from mangrove sediments in Guanabara Bay, Brazil. Res Microbiol 157:752–762
Bron S, Venema G (1972) Ultraviolet inactivation and excision-repair in Bacillus subtilis II. Differential inactivation and differential repair of transforming markers. Mutat Res Fund Mol Mech Mutagen 15:11–22
Cole J, Chai B, Marsh T, Farris R, Wang Q, Kulam S et al (2003) The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442–443
Eguchi M, Ostrowski M, Fegatella F, Bowman J, Nichols D, Nishino T et al (2001) Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl Environ Microbiol 67:4945–4954
Fegatella F, Lim J, Kjelleberg S, Cavicchioli R (1998) Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol 64:4433–4438
Filion M, St-Arnaud M, Jabaji-Hare SH (2003) Direct quantification of fungal DNA from soil substrate using real-time PCR. J Microbiol Methods 53:67–76
Guo C, Zhou H, Wong Y, Tam N (2005) Isolation of PAH-degrading bacteria from mangrove sediments and their biodegradation potential. Mar Pollut Bull 51:1054–1061
Ho Y, Jackson M, Yang Y, Mueller J, Pritchard P (2000) Characterization of fluoranthene-and pyrene-degrading bacteria isolated from PAH-contaminated soils and sediments. J Ind Microbiol Biotechnol 24:100–112
Jansson J, Leser T, Kowalchuk G, Bruijn F, Head I, Akkermans A et al (2004) Quantitative PCR of environmental samples. Molecular Microbial Ecology Manual 1, 2:445–463
Khan IUH, Yadav JS (2004) Real-time PCR assays for genus-specific detection and quantification of culturable and non-culturable mycobacteria and pseudomonads in metalworking fluids. Mol Cell Probe 18:67–73
Klappenbach JA, Saxman PR, Cole JR, Schmidt TM (2001) rrndb: the Ribosomal RNA Operon Copy Number database. Nucleic Acids Res 29:181–184
Laurel DC, Craig SC (2003) Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques 34:2–9
Leung K, Chang Y, Gan Y, Peacock A, Macnaughton S, Stephen J et al (1999) Detection of Sphingomonas spp in soil by PCR and sphingolipid biomarker analysis. J Ind Microbiol Biotechnol 23:252–260
Leys N, Ryngaert A, Bastiaens L, Verstraete W, Top E, Springael D (2004) Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl Environ Microbiol 70:1944–1955
Leys N, Ryngaert A, Bastiaens L, Top E, Verstraete W, Springael D (2005) Culture independent detection of Sphingomonas sp. EPA 505 related strains in soils contaminated with polycyclic aromatic hydrocarbons (PAHs). Microb Ecol 49:443–450
Li H, Zhang Y, Zhang C, Chen G (2005) Effect of petroleum-containing wastewater irrigation on bacterial diversities and enzymatic activities in a paddy soil irrigation area. J Environ Qual 34:1073–1080
Li H, Zhang Y, Kravchenko I, Xu H, Zhang C (2007) Dynamic changes in microbial activity and community structure during biodegradation of petroleum compounds: a laboratory experiment. J Environ Sci (China) 19:1003–1013
Li H, Zhang Y, Li D, Xu H, Chen G, Zhang C (2009) Comparisons of different hypervariable regions of rrs genes for fingerprinting of microbial communities in paddy soils. Soil Biol Biochem 41:954–968
Murakami Y, Otsuka S, Senoo K (2010) Abundance and community structure of Sphingomonads in leaf residues and nearby bulk soil. Microbes Environ 25:183–189
Mushinsky HR (1985) Fire and the Florida sandhill herpetofaunal community: with special attention to responses of Cnemidophorus sexlineatus. Herpetologica 41:333–342
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nakatsu C (2007) Soil microbial community analysis using denaturing gradient gel electrophoresis. Soil Sci Soc Am J 71:562–571
Padidam M, Beachy R, Fauquet C (1995) Classification and identification of geminiviruses using sequence comparisons. J Gen Virol 76:249–263
Pielou EC (1969) An introduction to mathematical ecology. Wiley, New York
Pinyakong O, Habe H, Omori T (2003) The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J Gen Appl Microbiol 49:1–19
Salazar O, Gonzalez I, Genilloud O (2002) New genus-specific primers for the PCR identification of novel isolates of the genera Nocardiopsis and Saccharothrix. Int J Syst Evol Microbiol 52:1411–1421
Saul D, Aislabie J, Brown C, Harris L, Foght J (2005) Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol Ecol 53:141–155
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Shi T, Fredrickson J, Balkwill D (2001) Biodegradation of polycyclic aromatic hydrocarbons by Sphingomonas strains isolated from the terrestrial subsurface. J Ind Microbiol Biotechnol 26:283–289
Shokrollahzadeh S, Azizmohseni F, Golmohammad F, Shokouhi H, Khademhaghighat F (2008) Biodegradation potential and bacterial diversity of a petrochemical wastewater treatment plant in Iran. Bioresource Technol 99:6127–6133
Skovhus TL, Ramsing NB, Holmstrom C, Kjelleberg S, Dahllof I (2004) Real-time quantitative PCR for assessment of abundance of Pseudoalteromonas species in marine samples. Appl Environ Microbiol 70:2373–2382
Ter Braak CJF, Smilauer P (2002) CANOCO reference manual and CanoDraw for Windows user's guide: Software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, NY, USA
Tiirola MA, Männistö MK, Puhakka JA, Kulomaa MS (2002) Isolation and characterization of Novosphingobium sp. strain MT1, a dominant polychlorophenol-degrading strain in groundwater bioremediation system. Appl Environ Microbiol 68:173–180
Vanbroekhoven K, Ryngaert A, Bastiaens L, Wattiau P, Vancanneyt M, Swings J et al (2004) Streptomycin as a selective agent to facilitate recovery and isolation of introduced and indigenous Sphingomonas from environmental samples. Environ Microbiol 6:1123–1136
Vitzthum F, Geiger G, Bisswanger H, Brunner H, Bernhagen J (1999) A quantitative fluorescence-based microplate assay for the determination of double-stranded DNA using SYBR Green I and a standard ultraviolet transilluminator gel imaging system. Anal Biochem 276:59–64
White DC, Sutton SD, Ringelberg DB (1996) The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol 7:301–306
Widmer F, Seidler R, Gillevet P, Watrud L, Di Giovanni G (1998) A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl Environ Microbiol 64:2545–2553
Wilsey B, Stirling G (2007) Species richness and evenness respond in a different manner to propagule density in developing prairie microcosm communities. Plant Ecol 190:259–273
Yabuuchi E, Yano I, Oyaizu H, Hashimoto Y, Ezaki T, Yamamoto H (1990) Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol 34:99–119
Yabuuchi E, Kosako Y, Fujiwara N, Naka T, Matsunaga I, Ogura H, Kobayashi K (2002) Emendation of the genus Sphingomonas Yabuuchi et al. 1990 and junior objective synonymy of the species of three genera, Sphingobium, Novosphingobium and Sphingopyxis, in conjunction with Blastomonas ursincola. Int J Syst Evol Microbiol 52:1485–1496
Acknowledgments
This work was supported by the National Nature Sciences Foundation of China No. 30800157 and the Knowledge Innovation Program of the Chinese Academy of Sciences, No. KSCX2-YW-G-071.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Elena Maestri
Rights and permissions
About this article
Cite this article
Zhou, L., Li, H., Zhang, Y. et al. Abundance and diversity of Sphingomonas in Shenfu petroleum-wastewater irrigation zone, China. Environ Sci Pollut Res 19, 282–294 (2012). https://doi.org/10.1007/s11356-011-0552-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-011-0552-y