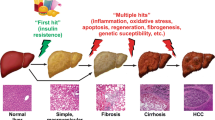
Abstract
Introduction
Untargeted metabolomics workflows include numerous points where variance and systematic errors can be introduced. Due to the diversity of the lipidome, manual peak picking and quantitation using molecule specific internal standards is unrealistic, and therefore quality peak picking algorithms and further feature processing and normalization algorithms are important. Subsequent normalization, data filtering, statistical analysis, and biological interpretation are simplified when quality data acquisition and feature processing are employed.
Objectives
Metrics for QC are important throughout the workflow. The robust workflow presented here provides techniques to ensure that QC checks are implemented throughout sample preparation, data acquisition, pre-processing, and analysis.
Methods
The untargeted lipidomics workflow includes sample standardization prior to acquisition, blocks of QC standards and blanks run at systematic intervals between randomized blocks of experimental data, blank feature filtering (BFF) to remove features not originating from the sample, and QC analysis of data acquisition and processing.
Results
The workflow was successfully applied to mouse liver samples, which were investigated to discern lipidomic changes throughout the development of nonalcoholic fatty liver disease (NAFLD). The workflow, including a novel filtering method, BFF, allows improved confidence in results and conclusions for lipidomic applications.
Conclusion
Using a mouse model developed for the study of the transition of NAFLD from an early stage known as simple steatosis, to the later stage, nonalcoholic steatohepatitis, in combination with our novel workflow, we have identified phosphatidylcholines, phosphatidylethanolamines, and triacylglycerols that may contribute to disease onset and/or progression.




Similar content being viewed by others
Abbreviations
- BFF:
-
Blank feature filtering
- Cer:
-
Ceramide
- DG:
-
Diacylglyceride
- LC:
-
Liquid chromatography
- MS:
-
Mass spectrometry
- NAFLD:
-
Nonalcoholic fatty liver disease
- NASH:
-
Nonalcoholic steatohepatitis
- QC:
-
Quality control
- RT:
-
Retention time
- SS:
-
Simple steatosis
- TG:
-
Triacylglyceride
References
Arendt, B. M., Ma, D. W., Simons, B., Noureldin, S. A., Therapondos, G., Guindi, M., et al. (2013). Nonalcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Applied Physiology, Nutrition, and Metabolism, 38(3), 334–340. doi:10.1139/apnm-2012-0261.
Bril, F., & Cusi, K. (2016). Nonalcoholic fatty liver disease: The new complication of type 2 diabetes mellitus. Endocrinology and Metabolism Clinics of North America, 45(4), 765–781. doi:10.1016/j.ecl.2016.06.005.
Broadhurst, D. I., & Kell, D. B. (2006). Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics, 2(4), 171–196. doi:10.1007/s11306-006-0037-z.
Browning, J. D., Szczepaniak, L. S., Dobbins, R., Nuremberg, P., Horton, J. D., Cohen, J. C., et al. (2004). Prevalence of hepatic steatosis in an urban population in the United States: Impact of ethnicity. Hepatology, 40(6), 1387–1395.
Castro-Perez, J. M., Kamphorst, J., Degroot, J., Lafeber, F., Goshawk, J., Yu, K., et al. (2010). Comprehensive LC–MS E lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients research articles. Journal of Proteome Research, 9, 2377–2389.
Chalasani, N., Younossi, Z., Lavine, J. E., Diehl, A. M., Brunt, E. M., Cusi, K., et al. (2012). The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology, 142(7), 1592–1609. doi:10.1053/j.gastro.2012.04.001.
Chambers, M. C., Maclean, B., Burke, R., Amodei, D., Ruderman, D. L., Neumann, S., et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotechnology, 30(10), 918–920. doi:10.1038/nbt.2377.
Cheng, D., Jenner, A. M., Shui, G., Cheong, W. F., Mitchell, T. W., Nealon, J. R., et al. (2011). Lipid pathway alterations in Parkinson’s disease primary visual cortex. PLoS ONE, 6(2), e17299. doi:10.1371/journal.pone.0017299.
Clapper, J. R., Hendricks, M. D., Gu, G., Wittmer, C., Dolman, C. S., Herich, J., et al. (2013). Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. American Journal of Physiology Gastrointestinal and Liver Physiology, 305(7), G483–G495. doi:10.1152/ajpgi.00079.2013.
De Livera, A. M., Dias, D. A., De Souza, D., Rupasinghe, T., Tull, D. L., Roessner, U., et al. (2012). Normalising and integrating metabolomics data. Analytical Chemistry, 84, 10768–10776
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 6(7), 1060–1083. doi:10.1038/nprot.2011.335.
Garrett, R. H., & Grisham, C. M. (2010). The tricarboxylic acid cycle. In L. Lockwood, S. Kiselica, A. Summers, & L. Weber (Eds.), Biochemistry (4th ed.). Boston: Cengage Learning.
Gibellini, F., & Smith, T. K. (2010). The Kennedy pathway-de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life, 62(6), 414–428. doi:10.1002/iub.337.
Gorden, D. L., Ivanova, P. T., Myers, D. S., McIntyre, J. O., VanSaun, M. N., Wright, J. K., et al. (2011). Increased diacylglycerols characterize hepatic lipid changes in progression of human nonalcoholic fatty liver disease; comparison to a murine model. PLoS ONE, 6(8), 1–10. doi:10.1371/journal.pone.0022775.
Gorden, D. L., Myers, D. S., Ivanova, P. T., Fahy, E., Maurya, M. R., Gupta, S., et al. (2015). Biomarkers of NAFLD progression: A lipidomics approach to an epidemic. Journal of Lipid Research, 56(3), 722–736. doi:10.1194/jlr.P056002.
Jacobs, R. L., van der Veen, J. N., & Vance, D. E. (2013). Finding the balance: The role of S-adenosylmethionine and phosphatidylcholine metabolism in development of nonalcoholic fatty liver disease. Hepatology (Baltimore, Md.), 58, 1207–1209. doi:10.1002/hep.26499.
Kirwan, J. A., Weber, R. J. M., Broadhurst, D. I., & Viant, M. R. (2014). Direct infusion mass spectrometry metabolomics dataset: A benchmark for data processing and quality control. Scientific Data, 1, 140012. doi:10.1038/sdata.2014.12.
Koelmel, J. P., Kroeger, N. M., Ulmer, C. Z., Bowden, J. A., Patterson, R. E., Cochran, J. A., et al. (2017). LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics, 18, 331. doi:10.1186/s12859-017-1744-3.
Koliaki, C., & Roden, M. (2013). Hepatic energy metabolism in human diabetes mellitus, obesity and non-alcoholic fatty liver disease. Molecular and Cellular Endocrinology, 379(1–2), 35–42. doi:10.1016/j.mce.2013.06.002.
Li, Z., Agellon, L. B., Allen, T. M., Umeda, M., Jewell, L., Mason, A., & Vance, D. E. (2006). The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metabolism, 3(5), 321–331. doi:10.1016/j.cmet.2006.03.007.
Martin Bland, J., & Altman, D. (1986). Statistical methods for assessing agreement between two methods of clinical measurement. Lancet, 327(8476), 307–310. doi:10.1016/S0140-6736(86)90837-8.
Nagarajan, P. (2012). Genetically modified mouse models for the study of nonalcoholic fatty liver disease. World Journal of Gastroenterology, 18(11), 1141. doi:10.3748/wjg.v18.i11.1141.
Patterson, R. E., Kalavalapalli, S., Williams, C. M., Nautiyal, M., Mathew, J. T., Martinez, J., et al. (2016). Lipotoxicity in steatohepatitis occurs despite an increase in tricarboxylic acid cycle activity. American Journal of Physiology—Endocrinology and Metabolism, 310(7), E484–E494. doi:10.1152/ajpendo.00492.2015.
Pellicoro, A., Ramachandran, P., Iredale, J. P., & Fallowfield, J. A. (2014). Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nature Reviews Immunology, 14(3), 181–194. doi:10.1038/nri3623.
Pluskal, T., Castillo, S., Villar-Briones, A., & Orešič, M. (2010). MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics, 11(1), 395. doi:10.1186/1471-2105-11-395.
Qi, Z., & Voit, E. O. (2016). Strategies for comparing metabolic profiles: Implications for the inference of biochemical mechanisms from metabolomics data. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 99, 1–32. doi:10.1109/TCBB.2016.2586065.
Redestig, H., Fukushima, A., Stenlund, H., Moritz, T., Arita, M., Saito, K., & Kusano, M. (2009). Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Analytical Chemistry, 81(19), 7974–7980. doi:10.1021/ac901143w.
Satapati, S., Sunny, N. E., Kucejova, B., Fu, X., He, T. T., Méndez-Lucas, A., et al. (2012). Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. Journal of Lipid Research, 53(6), 1080–1092. doi:10.1194/jlr.M023382.
Silva, L. P., Lorenzi, P. L., Purwaha, P., Yong, V., Hawke, D. H., & Weinstein, J. N. (2014). Measurement of DNA concentration as a normalization strategy for metabolomic data from adherent cell lines. Analytical Chemistry, 85(20), 9536–9542. doi:10.1021/ac401559v.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. a., et al. (2007). Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics, 3(3), 211–221. doi:10.1007/s11306-007-0082-2.
Sunny, N. E., Bril, F., & Cusi, K. (2016). Mitochondrial adaptation in nonalcoholic fatty liver disease: Novel mechanisms and treatment strategies. Trends in Endocrinology & Metabolism, 28, 1–11. doi:10.1016/j.tem.2016.11.006.
Sunny, N. E., Parks, E. J., Browning, J. D., & Burgess, S. C. (2011). Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metabolism, 14(6), 804–810. doi:10.1016/j.cmet.2011.11.004.
Trevaskis, J. L., Griffin, P. S., Wittmer, C., Neuschwander-Tetri, B. A., Brunt, E. M., Dolman, C. S., et al. (2012). Glucagon-like peptide-1 receptor agonism improves metabolic, biochemical, and histopathological indices of nonalcoholic steatohepatitis in mice. American Journal of Physiology Gastrointestinal and Liver Physiology, 302(38), G762–G772. doi:10.1152/ajpgi.00476.2011.
Vorkas, P. A., Isaac, G., Anwar, M. A., Davies, A. H., Want, E. J., Nicholson, J. K., & Holmes, E. (2015). Untargeted UPLC-MS profiling pipeline to expand tissue metabolome coverage: Application to cardiovascular disease. Analytical Chemistry, 87(8), 4184–4193. doi:10.1021/ac503775m.
Worley, B., & Powers, R. (2013). Multivariate analysis in metabolomics. Current Metabolomics, 1, 92–107.
Acknowledgements
The authors would like to thank Miguel Ibarra, Oleksandr Moskalenko, and Justin Richardson for their computer programming expertise. We would also like to thank funding sources including the Southeastern Center for Integrated Metabolomics (NIH grant U24 DK097209), the UF Clinical Translational Science Institute (CTSI NIH Grant UL1 TR000064), and the Eastman Chemical Company Analytical Summer Fellowship.
Funding
This study was funded by NIH grant U24 DK097209, UF Clinical Translational Science Institute (CTSI NIH Grant UL1 TR000064).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patterson, R.E., Kirpich, A.S., Koelmel, J.P. et al. Improved experimental data processing for UHPLC–HRMS/MS lipidomics applied to nonalcoholic fatty liver disease. Metabolomics 13, 142 (2017). https://doi.org/10.1007/s11306-017-1280-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1280-1