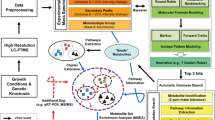
Abstract
Gene set enrichment analysis (GSEA) has been successfully employed in transcriptomics and proteomics research for over 5 years. We have applied a modified GSEA approach to metabolomics, called metabolite set enrichment analysis (MSEA), and have integrated this method into the MeltDB platform. With the novel integrated functionality, we evaluate the applicability of the approach for metabolomics research by analyzing the metabolic profiles of industrial amino acid producer strains of Corynebacterium glutamicum. Metabolite sets are obtained from metabolic pathways defined in the KEGG and the newly created CglCyc databases. In the first experiment using MSEA, the metabolic profiling analyses compared glucose- and acetate-grown strains, and, in the second, a production strain series in which the carbon flow is shifted step-wise from the lysine pathway to the branching threonine, isoleucine, and methionine pathways. By identifying changes of the metabolic profile MSEA was able to identify the metabolic pathways activated while utilizing different carbon sources or those that have been genetically modified. The presented experimental results are publicly available at http://meltdb.cebitec.uni-bielefeld.de.



Similar content being viewed by others
References
Aggio, R. B., Ruggiero, K., & Villas-Boas, S. G. (2010). Pathway activity profiling (PAPi): From the metabolite profile to the metabolic pathway activity. Bioinformatics, 26(23), 2969–2976.
Auchter, M., Arndt, A., & Eikmanns, B. J. (2009). Dual transcriptional control of the acetaldehyde dehydrogenase gene ald of Corynebacterium glutamicum by RamA and RamB. Journal of Biotechnology, 140(1–2), 84–91.
Becker, J., Klopprogge, C., Schroder, H., & Wittmann, C. (2009). Metabolic engineering of the tricarboxylic acid cycle for improved lysine production by Corynebacterium glutamicum. Applied and Environmental Microbiology, 75(24), 7866–7869.
Becker, J., Klopprogge, C., & Wittmann, C. (2008). Metabolic responses to pyruvate kinase deletion in lysine producing Corynebacterium glutamicum. Microbial Cell Factories, 7, 8.
Blombach, B., & Seibold, G. M. (2010). Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Applied Microbiology and Biotechnology, 86(5), 1313–1322.
Born, T. L., & Blanchard, J. S. (1999). Enzyme-catalyzed acylation of homoserine: Mechanistic characterization of the Escherichia coli metA-encoded homoserine transsuccinylase. Biochemistry, 38(43), 14416–14423.
Burkovski, A. (2006). Proteomics of Corynebacterium glutamicum: Essential industrial bacterium. Methods of Biochemical Analysis, 49, 137–147.
de Koning, W., & van Dam, K. (1992). A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Analytical Biochemistry, 204(1), 118–123.
Drysch, A., El Massaoudi, M., Mack, C., et al. (2003). Production process monitoring by serial mapping of microbial carbon flux distributions using a novel sensor reactor approach: II–(13)C-labeling-based metabolic flux analysis and l-lysine production. Metabolic Engineering, 5(2), 96–107.
Drysch, A., El Massaoudi, M., Wiechert, W., de Graaf, A. A., & Takors, R. (2004). Serial flux mapping of Corynebacterium glutamicum during fed-batch l-lysine production using the sensor reactor approach. Biotechnology and Bioengineering, 85(5), 497–505.
Dunn, W. B., & Ellis, D. I. (2005). Metabolomics: Current analytical platforms and methodologies. Trends in Analytical Chemistry, 4, 285–294.
El Massaoudi, M., Spelthahn, J., Drysch, A., de Graaf, A., & Takors, R. (2003). Production process monitoring by serial mapping of microbial carbon flux distributions using a novel sensor reactor approach: I–Sensor reactor system. Metabolic Engineering, 5(2), 86–95.
Fernie, A. R., Trethewey, R. N., Krotzky, A. J., & Willmitzer, L. (2004). Metabolite profiling: From diagnostics to systems biology. Nature Reviews. Molecular Cell Biology, 5(9), 763–769.
Fiehn, O. (2002). Metabolomics—the link between genotypes and phenotypes. Plant Molecular Biology, 48(1–2), 155–171.
Gerstmeir, R., Wendisch, V. F., Schnicke, S., et al. (2003). Acetate metabolism and its regulation in Corynebacterium glutamicum. Journal of Biotechnology, 104(1–3), 99–122.
Goodacre, R., Vaidyanathan, S., Dunn, W. B., Harrigan, G. G., & Kell, D. B. (2004). Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends in Biotechnology, 22(5), 245–252.
Guillouet, S., Rodal, A. A., An, G. H., et al. (2001). Metabolic redirection of carbon flow toward isoleucine by expressing a catabolic threonine dehydratase in a threonine-overproducing Corynebacterium glutamicum. Applied Microbiology and Biotechnology, 57(5–6), 667–673.
Hall, R. D. (2006). Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytologist, 169(3), 453–468.
Han, S. O., Inui, M., & Yukawa, H. (2008). Transcription of Corynebacterium glutamicum genes involved in tricarboxylic acid cycle and glyoxylate cycle. Journal of Molecular Microbiology and Biotechnology, 15(4), 264–276.
Hansmeier, N., Chao, T. C., Puhler, A., Tauch, A., & Kalinowski, J. (2006). The cytosolic, cell surface and extracellular proteomes of the biotechnologically important soil bacterium Corynebacterium efficiens YS-314 in comparison to those of Corynebacterium glutamicum ATCC 13032. Proteomics, 6(1), 233–250.
Hayashi, M., Mizoguchi, H., Shiraishi, N., et al. (2002). Transcriptome analysis of acetate metabolism in Corynebacterium glutamicum using a newly developed metabolic array. Bioscience, Biotechnology, and Biochemistry, 66(6), 1337–1344.
Hermann, T. (2003). Industrial production of amino acids by coryneform bacteria. Journal of Biotechnology, 104(1–3), 155–172.
Hoon Yang, T., Wittmann, C., & Heinzle, E. (2006). Respirometric 13C flux analysis–Part II: In vivo flux estimation of lysine-producing Corynebacterium glutamicum. Metabolic Engineering, 8(5), 432–446.
Huang, D. W., Sherman, B. T., & Lempicki, R. A. (2009). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research, 37(1), 1–13.
Hüser, A. T., Becker, A., Brune, I., et al. (2003). Development of a Corynebacterium glutamicum DNA microarray and validation by genome-wide expression profiling during growth with propionate as carbon source. Journal of Biotechnology, 106(2–3), 269–286.
Hüser, A. T., Chassagnole, C., Lindley, N. D., et al. (2005). Rational design of a Corynebacterium glutamicum pantothenate production strain and its characterization by metabolic flux analysis and genome-wide transcriptional profiling. Applied and Environmental Microbiology, 71(6), 3255–3268.
Hwang, B. J., Kim, Y., Kim, H. B., et al. (1999). Analysis of Corynebacterium glutamicum methionine biosynthetic pathway: Isolation and analysis of metB encoding cystathionine gamma-synthase. Molecules and Cells, 9(3), 300–308.
Hwang, B. J., Yeom, H. J., Kim, Y., & Lee, H. S. (2002). Corynebacterium glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. Journal of Bacteriology, 184(5), 1277–1286.
Ikeda, M., Ohnishi, J., Hayashi, M., & Mitsuhashi, S. (2006). A genome-based approach to create a minimally mutated Corynebacterium glutamicum strain for efficient l-lysine production. Journal of Industrial Microbiology & Biotechnology, 33(7), 610–615.
Kalinowski, J., Bathe, B., Bartels, D., et al. (2003). The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. Journal of Biotechnology, 104(1–3), 5–25.
Kanehisa, M., Goto, S., Hattori, M. et al. (2006). From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Research, 34(Database issue), D354–D357.
Karp, P. D., Paley, S. M., Krummenacker, M., et al. (2010). Pathway Tools version 13.0: Integrated software for pathway/genome informatics and systems biology. Briefings in Bioinformatics, 11(1), 40–79.
Kawaguchi, H., Sasaki, M., Vertes, A. A., Inui, M., & Yukawa, H. (2009). Identification and functional analysis of the gene cluster for l-arabinose utilization in Corynebacterium glutamicum. Applied and Environmental Microbiology, 75(11), 3419–3429.
Keilhauer, C., Eggeling, L., & Sahm, H. (1993). Isoleucine synthesis in Corynebacterium glutamicum: Molecular analysis of the ilvB-ilvN-ilvC operon. Journal of Bacteriology, 175(17), 5595–5603.
Kiefer, P., Heinzle, E., & Wittmann, C. (2002). Influence of glucose, fructose and sucrose as carbon sources on kinetics and stoichiometry of lysine production by Corynebacterium glutamicum. Journal of Industrial Microbiology & Biotechnology, 28(6), 338–343.
Krömer, J. O., Fritz, M., Heinzle, E., & Wittmann, C. (2005). In vivo quantification of intracellular amino acids and intermediates of the methionine pathway in Corynebacterium glutamicum. Analytical Biochemistry, 340(1), 171–173.
Krömer, J. O., Sorgenfrei, O., Klopprogge, K., Heinzle, E., & Wittmann, C. (2004). In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. Journal of Bacteriology, 186(6), 1769–1784.
Lee, H. S., & Hwang, B. J. (2003). Methionine biosynthesis and its regulation in Corynebacterium glutamicum: Parallel pathways of transsulfuration and direct sulfhydrylation. Applied Microbiology and Biotechnology, 62(5–6), 459–467.
Leuchtenberger, W. (1996) Amino acids: Technical production and use. In H. J. Rehm, G. Reed, A. Pühler, & P. Stadler (Eds.), Biotechnology (pp. 466–502). Weinheim: VCH.
Leuchtenberger, W., Huthmacher, K., & Drauz, K. (2005). Biotechnological production of amino acids and derivatives: Current status and prospects. Applied Microbiology and Biotechnology, 69(1), 1–8.
Lu, S. C. (2000). S-Adenosylmethionine. International Journal of Biochemistry and Cell Biology, 32(4), 391–395.
Mootha, V. K., Lindgren, C. M., Eriksson, K.-F., et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics, 34(3), 267–273.
Morbach, S., Sahm, H., & Eggeling, L. (1996). l-Isoleucine production with Corynebacterium glutamicum: Further flux increase and limitation of export. Applied and Environmental Microbiology, 62(12), 4345–4351.
Muffler, A., Bettermann, S., Haushalter, M., et al. (2002). Genome-wide transcription profiling of Corynebacterium glutamicum after heat shock and during growth on acetate and glucose. Journal of Biotechnology, 98(2–3), 255–268.
Neuweger, H., Albaum, S. P., Dondrup, M., et al. (2008). MeltDB: A software platform for the analysis and integration of metabolomics experiment data. Bioinformatics, 24(23), 2726–2732.
Neuweger, H., Persicke, M., Albaum, S. P., et al. (2009). Visualizing post genomics data-sets on customized pathway maps by ProMeTra-aeration-dependent gene expression and metabolism of Corynebacterium glutamicum as an example. BMC Systems Biology, 3, 82.
Nobeli, I., Ponstingl, H., Krissinel, E. B., & Thornton, J. M. (2003). A structure-based anatomy of the E. coli metabolome. Journal of Molecular Biology, 334(4), 697–719.
Nöh, K., Gronke, K., Luo, B., et al. (2007). Metabolic flux analysis at ultra short time scale: Isotopically non-stationary 13C labeling experiments. Journal of Biotechnology, 129(2), 249–267.
Ohnishi, J., Mitsuhashi, S., Hayashi, M., et al. (2002). A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine-producing mutant. Applied Microbiology and Biotechnology, 58(2), 217–223.
Parche, S., Burkovski, A., Sprenger, G. A., et al. (2001). Corynebacterium glutamicum: A dissection of the PTS. Journal of Molecular Microbiology and Biotechnology, 3(3), 423–428.
Park, S. D., Lee, J. Y., Kim, Y., Kim, J. H., & Lee, H. S. (1998). Isolation and analysis of metA, a methionine biosynthetic gene encoding homoserine acetyltransferase in Corynebacterium glutamicum. Molecules and Cells, 8(3), 286–294.
Park, S. D., Lee, J. Y., Sim, S. Y., Kim, Y., & Lee, H. S. (2007). Characteristics of methionine production by an engineered Corynebacterium glutamicum strain. Metabolic Engineering, 9(4), 327–336.
Persicke, M., Plassmeier, J., Neuweger, H. et al. (2010). Size exclusion chromatography-an improved method to harvest Corynebacterium glutamicum cells for the analysis of cytosolic metabolites. Journal of Biotechnology. doi:10.1016/j.jbiotec.2010.08.016.
Plassmeier, J., Barsch, A., Persicke, M., Niehaus, K., & Kalinowski, J. (2007). Investigation of central carbon metabolism and the 2-methylcitrate cycle in Corynebacterium glutamicum by metabolic profiling using gas chromatography-mass spectrometry. Journal of Biotechnology, 130(4), 354–363.
Rey, D. A., Nentwich, S. S., Koch, D. J., et al. (2005). The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Molecular Microbiology, 56(4), 871–887.
Rey, D. A., Pühler, A., & Kalinowski, J. (2003). The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. Journal of Biotechnology, 103(1), 51–65.
Rubin, E. (2006). Circumventing the cut-off for enrichment analysis. Briefings in Bioinformatics, 7(2), 202–203.
Rückert, C., Koch, D. J., Rey, D. A., et al. (2005). Functional genomics and expression analysis of the Corynebacterium glutamicum fpr2-cysIXHDNYZ gene cluster involved in assimilatory sulphate reduction. BMC Genomics, 6, 121.
Rückert, C., Pühler, A., & Kalinowski, J. (2003). Genome-wide analysis of the l-methionine biosynthetic pathway in Corynebacterium glutamicum by targeted gene deletion and homologous complementation. Journal of Biotechnology, 104(1–3), 213–228.
Schäfer, U., Boos, W., Takors, R., & Weuster-Botz, D. (1999). Automated sampling device for monitoring intracellular metabolite dynamics. Analytical Biochemistry, 270(1), 88–96.
Seibold, G., Auchter, M., Berens, S., Kalinowski, J., & Eikmanns, B. J. (2006). Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: Growth and lysine production. Journal of Biotechnology, 124(2), 381–391.
Seibold, G. M., Wurst, M., & Eikmanns, B. J. (2009). Roles of maltodextrin and glycogen phosphorylases in maltose utilization and glycogen metabolism in Corynebacterium glutamicum. Microbiology, 155(Pt 2), 347–358.
Stavrum, A. K., Petersen, K., Jonassen, I., & Dysvik, B. (2008). Analysis of gene-expression data using J-Express. Current Protocols in Bioinformatics, Chap. 7, Unit 7.3.
Subramanian, A., Kuehn, H., Gould, J., Tamayo, P., & Mesirov, J. P. (2007). GSEA-P: A desktop application for gene set enrichment analysis. Bioinformatics, 23(23), 3251–3253.
Subramanian, A., Tamayo, P., Mootha, V. K., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America, 102(43), 15545–15550.
Tauch, A., Götker, S., Pühler, A., Kalinowski, J., & Thierbach, G. (2002). The alanine racemase gene alr is an alternative to antibiotic resistance genes in cloning systems for industrial Corynebacterium glutamicum strains. Journal of Biotechnology, 99(1), 79–91.
van den Berg, R. A., Hoefsloot, H. C., Westerhuis, J. A., Smilde, A. K., & van der Werf, M. J. (2006). Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics, 7, 142.
Veit, A., Rittmann, D., Georgi, T., et al. (2009). Pathway identification combining metabolic flux and functional genomics analyses: acetate and propionate activation by Corynebacterium glutamicum. Journal of Biotechnology, 140(1–2), 75–83.
Wendisch, V. F., Bott, M., Kalinowski, J., Oldiges, M., & Wiechert, W. (2006). Emerging Corynebacterium glutamicum systems biology. Journal of Biotechnology, 124(1), 74–92.
Wendisch, V. F., de Graaf, A. A., Sahm, H., & Eikmanns, B. J. (2000). Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. Journal of Bacteriology, 182(11), 3088–3096.
Wittmann, C., Kiefer, P., & Zelder, O. (2004). Metabolic fluxes in Corynebacterium glutamicum during lysine production with sucrose as carbon source. Applied and Environmental Microbiology, 70(12), 7277–7287.
Xia, J., & Wishart, D. S. (2010). MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Research, 38(Suppl), W71–W77.
Acknowledgments
HN and LJS acknowledge the financial support of the SysLogics Project (BMBF 0315275A) and MP that of the SysMAP Project (BMBF 0313704). NK acknowledges the support by a fellowship from the CLIB-Graduate Cluster Industrial Biotechnology. The authors wish to thank the BRF team for expert technical support. Furthermore, the authors thank Daniel Stephen Brooks (Department of Philosophy, University of Bielefeld, Germany) and John Quimby (Massachusetts Institute of Technology, Sinskey Laboratory, Cambridge, USA) for carefully reading the manuscript and helping with wording and phrasing.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11306_2011_311_MOESM1_ESM.tif
Suppl. Figure 1: The visualization of the Enrichment Analysis results generated out of SVG graphics from the web interface. The best five results for the CglCyc metabolic pathways (A) exhibiting the highest enrichment scores were presented. The running ES score is plotted using blue color if the metabolites belonging to MS are predominantly found in the top part of the ordered List L. The red color indicates that the metabolites are found at the lower part of L. All metabolites present in the experiment are represented as gray boxes and the ones belonging to MS are labeled and highlighted in green. On the right, the computed m-values (B), the t-test probabilities, and the signal to noise ratios (SNR) are listed for all compounds identified in the analyzed metabolomics experiment of C. glutamicum DM1730 in comparison to the wild type. For the background of the table cells, a color mapping function of the m-values from red to green was chosen. (TIFF 35,835 kb)
11306_2011_311_MOESM2_ESM.tif
Suppl. Figure 2: The visualization of the Enrichment Analysis results generated out of SVG graphics from the web interface. The best five results for the CglCyc metabolic pathways (A) exhibiting the highest enrichment scores were presented. The running ES score is plotted using blue color if the metabolites belonging to MS are predominantly found in the top part of the ordered List L. The red color indicates that the metabolites are found at the lower part of L. All metabolites present in the experiment are represented as gray boxes and the ones belonging to MS are labeled and highlighted in green. On the right, the computed m-values (B), the t-test probabilities, and the signal to noise ratios (SNR) are listed for all compounds identified in the analyzed metabolomics experiment of C. glutamicum MP001 in comparison to the wild type. For the background of the table cells, a color mapping function of the m-values from red to green was chosen. (TIFF 35,119 kb)
11306_2011_311_MOESM3_ESM.tif
Suppl. Figure 3: The visualization of the Enrichment Analysis results generated out of SVG graphics from the web interface. The best five results for the CglCyc metabolic pathways (A) exhibiting the highest enrichment scores were presented. The running ES score is plotted using blue color if the metabolites belonging to MS are predominantly found in the top part of the ordered List L. The red color indicates that the metabolites are found at the lower part of L. All metabolites present in the experiment are represented as gray boxes and the ones belonging to MS are labeled and highlighted in green. On the right, the computed m-values (B), the t-test probabilities, and the signal to noise ratios (SNR) are listed for all compounds identified in the analyzed metabolomics experiment of C. glutamicum DM1795 in comparison to the wild type. For the background of the table cells, a color mapping function of the m-values from red to green was chosen. (TIFF 35,261 kb)
Rights and permissions
About this article
Cite this article
Persicke, M., Rückert, C., Plassmeier, J. et al. MSEA: metabolite set enrichment analysis in the MeltDB metabolomics software platform: metabolic profiling of Corynebacterium glutamicum as an example. Metabolomics 8, 310–322 (2012). https://doi.org/10.1007/s11306-011-0311-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-011-0311-6