Abstract
A metabolomics-based systems toxicology approach was used to profile the urinary metabolites for the toxicity related processes and pathogenesis induced by doxorubicin (DOX) to rats. Endogenous metabolite profiles were obtained with ultra performance liquid chromatography-mass spectrometry (UPLC-MS) for rats receiving different single dosages of DOX (5, 10 or 20 mg/kg) prior and at three time points after dosage. Principal components analysis (PCA) allowed detection of two major systemic metabolic changes with the time due to the induced toxicity. Furthermore, Analysis of variance (ANOVA) Simultaneous Component Analysis (ASCA) was applied to reveal the variation caused by time and dose, and their interaction in a multivariate way. Finally, various metabolites involved in the toxic processes could be identified using their accurate mass and MSn experiments, and possible mechanisms of the toxicity of DOX were postulated. In conclusion, metabolomics as a systems toxicology approach was able to provide comprehensive information on the dynamic process of drug induced toxicity. In addition, detection of the systemic toxic effects could be obtained with metabolomics at an earlier stage compared to the clinical chemistry and histopathological assessment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The rapid improvements of the technologies enabling the determination of large numbers of biomolecules and subsequent multivariate data analysis provide the opportunity to obtain new insights in the toxicity by a biochemically based systems toxicology approach (Van der Greef et al. 2007; Van der Greef et al. 2004). In such an approach, the biochemical changes in preferably easily accessible biological fluids such as blood or urine are detected as biomarkers for toxicity-related pathogenesis. The biomarkers can be mapped onto known biochemical pathways, and may be then confirmed to correspond to toxic compound related biochemical processes, or may suggest new biochemical pathways. Moreover, some biomarkers may be indirect markers and can only be understood in the perspective of the whole system, offering the potential opportunity to explore novel biochemistry and reveal new insights in the toxicity mechanisms. In the context of systems biology, metabolomics acquires signals representing tens to hundreds of metabolites in biofluids and enables assessment of toxicity in vivo, providing rapid, often minimal invasive, robust and reproducible information about biochemical changes due to toxicity and has shown particular promise in the area of toxicology as a systems toxicity approach (Nicholson et al. 1999; Keun 2006; Waters et al. 2005). Extensive research has been performed by using Nuclear Magnetic Resonance (NMR) to investigate the mechanisms, site and severity of toxic effects of model toxins (Lindon et al. 2003). Because of the high sensitivity, electrospray mass spectrometry, especially coupled with liquid chromatography, has emerged as a complementary technique for generating metabolomics data and has been applied to different biofluids (urine, plasma, synovial fluid) and a wide scope of metabolites (Kamphorst et al. 2007; Lafaye et al. 2003; Yang et al. 2006; Lenz et al. 2004; Williams et al. 2005). Ultra performance liquid chromatography (UPLC) employs porous particles with internal diameters smaller than 2 μm, resulting in higher peak capacity, improved resolution and increased sensitivity compared to conventional HPLC columns, therefore making it even more suitable for a metabolomics approach (Theodoridis et al. 2008).
Doxorubicin (DOX, Fig. 1e) is a topoisomerase II poison and widely used in the treatment of various human neoplasms (Singal and Iliskovic 1998; De Beer et al. 2001; Swift et al. 2006). However, the clinical use of DOX is limited by the development of a dose-dependent cumulative cardiotoxicity (Singal and Iliskovic 1998; De Beer et al. 2001). Furthermore, animal studies reveal potential liver and kidney lesions. It was reported that DOX could induce hepatic lipid peroxidation (Bagchi et al. 1995) and changes in the kidney of rats including increases of glomerular capillary permeability and tubular atrophy (Wapstra et al. 1999; Saad et al. 2001). The exact causal mechanisms of DOX induced toxicity remain unclear and various mechanisms have been proposed to interpret the toxicity, including inhibition of nucleic acid and protein synthesis (Meliksetian et al. 1997), direct interaction with the actin–myosin contractile system, anthracycline metabolite hypothesis, alterations in platelet-activating factor, prostaglandin and intracellular calcium etc. (De Beer et al. 2001). Most of the evidences indicate that free radicals are involved (De Beer et al. 2001; Kalender et al. 2005; Oliveira et al. 2004). In this study, we applied a non-invasive, sensitive UPLC-MS based urine metabolite profile approach to investigate the lesions induced by DOX. The purpose was to investigate the systemic toxic processes of DOX induced toxicities including onset and progression, and the potential mechanisms involved. For this, pattern-recognition algorithms principal components analysis (PCA) and ANOVA simultaneous component analysis (ASCA) were used to find systemic metabolic changes with time, dose and their interaction. In addition, the identity of potential biomarkers found had to be revealed to allow interpretation.
Full-scan UPLC-MS metabolite profiles of urine samples of the same rat of the middle dose a 24 h before administration and b 24, c 48 and d 96 h after administration of DOX. The inset e shows the structure of DOX, which is detected in urine after administration at 14.7 min. The inset f shows a typical mass spectrum of the chromatographic peak with a retention time of 16.7 min. The inset g is the enlarged chromatogram from 0 to 14 min of d. All chromatograms are normalized on the intensity of the highest peak
2 Methods
2.1 Chemicals
Doxorubicin hydrochloride was obtained from ShangHai Hualian Pharmaceuticals (Shanghai, China; batch No. 040502), and diluted with 0.9% (w/v) saline to a concentration of 1.25 mg/ml, 2.5 mg/ml and 5 mg/ml for intraperitoneal injection. Chemical standards such as cholic acid, hippuric acid, uric acid and 2,8-quinolinediol were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile, HPLC grade, was purchased from Fisher Scientific (Loughborough, UK). Formic acid (HPLC/SPECTRO grade) was purchased from Tedia (Fairfield, OH, USA). Water was produced by a Milli-Q Ultra-pure water system (Millipore, Billerica, USA). Leucine-enkephalin and β-glucuronidase were obtained from Sigma-Aldrich (St. Louis, MO, USA).
2.2 Animals treatments and samples collection
Male Wistar rats weighing 280–320 g were obtained from Beijing National Medicine Evaluation Center (Beijing, China). The rats were housed under controlled humidity (35–50%) and temperature (20–25°C) with a 12 h light/dark cycle. Animals had free access to certified rodent chow (National Medicine Evaluation Center, Beijing, China) and water.
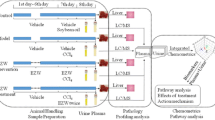
For metabolomics, a total of 20 rats were randomly divided into four groups with five rats in each group, i.e. control, low, middle and high dose group, and housed in metabolism cages. The rats of low, middle and high dose group received a single dose of 5 mg/kg, 10 mg/kg and 20 mg/kg DOX intraperitoneally. The control group received a single dose of the same volume of 0.9% (w/v) saline. Urine samples were collected over ice during 24 h before administration (−24 h) and 0–24 h, 24–48 h and 72–96 h after dosing. Urine samples were centrifuged (3,000×g, 10 min) and the supernatants were stored at −80°C until analysis.
For clinical chemistry and histopathology, 60 rats were grouped and dosed as above (15 animals per group). Five rats were selected from each group randomly and sacrificed at 24, 48 and 96 h after administration. The plasma samples were analyzed by standard clinical chemistry methods using a Hitachi 7020 Automatic Analyzer (Hitachi, Tokyo, Japan) for aspartate aminotransferase (AST), alanine aminotransferase (ALT), creatine kinase (CK), creatinine (CREA), blood urea nitrogen (BUN) and lactate dehydrogenase (LDH) (Saad et al. 2001). Significant changes compared to the control group in these parameters were determined using a Student’s t-test (Microsoft Excel version 2002). The target organs, i.e. heart, liver and kidney, were processed for paraffin sectioning, stained and examined using light microscopy.
2.3 Sample preparation
Prior to analysis, the urine samples were thawed at room temperature and centrifuged under 10,000g for 5 min. The supernatant was diluted with water in the ratio of 1:4 (v/v).
2.4 Chromatography
Chromatographic separations were performed on a 100 × 2.1 mm ACQUITY-1.7 μm C18 column (Waters Corp, Milford, USA) using an ACQUITY-Ultra Performance Liquid Chromatography system (Waters). A “purge-wash-purge” cycle was employed on the autosampler, with 80% aqueous methanol as the wash solvent and 10% aqueous methanol as the purge solvent to ensure that the carry-over between injections was minimized.
The column was maintained at 35°C and a gradient of 0.1% aqueous formic acid (solvent A) and acetonitrile (solvent B) used as follows: a linear gradient from 0% to 25% solvent B in 14 min, subsequently a linear gradient from 25% to 100% B in 4 min, and then 100% B kept for 1.5 min. Next, the solvent was returned to 100% A within 0.1 min and the system equilibrated for 1.5 min. The flow rate was 0.35 ml/min and a 5 μl aliquot of each sample was injected onto the column. Each sample was analyzed in duplicate. The eluent was introduced to the mass spectrometry directly, i.e. without a split.
2.5 Mass spectrometry
Mass spectrometry was performed on a Micromass oa-Q-Tof (Waters MS Technologies, Manchester, UK). The desolvation gas was set to 500 l/h at a temperature of 350°C, the cone gas to 50 l/h and the source temperature to 100°C. The capillary voltage was set at 2,900 V in positive ion mode and 2,600 V in negative ion mode, and the cone voltage was set at 35 V. The data acquisition rate was set to 0.4 s/scan, with a 0.1 s interscan delay. For accurate mass acquisition, a lock-mass of leucine enkephalin at a concentration of 2 ng/μl in methanol/1% aqueous acetic acid (50/50 v/v) was used via a lock spray interface at a flow rate of 10 μl/min monitoring for positive ion mode ([M + H]+ = 556.2771) and negative mode([M − H]− = 554.2615). Data were collected in centroid mode from 50 m/z to 850 m/z with a lockspray acquisition of 1 s every 20 s, and the lock mass data were averaged over 10 scans for correction.
2.6 Data processing, multivariate analysis and biomarker identification
The LC-MS data were analyzed using the Micromass MarkerLynx Applications Manager (Version 4.0, Waters, UK). An ApexTrack- peak detection algorithm was used in MarkerLynx to detect peaks and align retention times of the peaks for all chromatograms. The data were combined into a single data matrix by aligning peaks with the same mass/retention time pair together from each data file in the dataset, along with their associated intensities. The differences in concentration of compounds between individual urine samples are compensated by peak normalization. The intensities of the peaks were normalized so that the sum of the intensities of all peaks of a single LC-MS run were equal between different runs. The intensities of each peak of two duplicate injections were averaged and the final dataset consisted of 80 (20 × 4) samples. Peaks derived from DOX and its possible metabolites were excluded from the dataset. DOX in urine samples was excluded by the comparison of retention time and MS spectrum of DOX chemical standard. If a peak was only detected in the samples after dose, then metabolynx (Version 4.0, Waters, U.K.) was used to find if it was a possible metabolite of DOX by the differences in mass that can be attributed to metabolic transformation such as loss of methyl, addition of oxygen or a conjugate. With above described method, two datasets were constructed, the first dataset included the peaks from 0.5 min to 14 min and the second dataset included the peaks from 0.5 min to 18 min.
The resulting dataset comprising peak numbers (RT—m/z pair), sample names, and ion intensities was analyzed by PCA and ASCA in MATLAB (version R2006b, The Mathworks, Natick, MA).
Identification of potential biomarkers was based on online and off-line LC-MS techniques. The accurate mass determinations was performed by online UPLC-oa-Q-Tof with lockspray or by fraction collection of the LC eluent and determined with a linear-ion trap-Fourier-transform ion cyclotron resonance mass spectrometer (LTQ-FTMS, Thermo Fisher, San Jose, CA, USA) by direct infusion experiments. MS2 spectra were generated with Micromass oa-Q-Tof and MS3 spectra were generated with QSTAR (Pulsar i, PE/Sciex, Canada) and LTQ-FTMS for identification of 2,8-quinolinediol glucuronic acid conjugate. Finally, the provisionally identifications of metabolites were validated with authentic chemical standards by comparison of the retention times and MS/MS spectra. The identification of the glucuronic acid conjugate involved enzyme degradation. The appropriate fraction of the LC eluent containing glucuronic acid conjugate was freeze dried and subsequently incubated with β-glucuronidase solution for 12 h (pH 4.7, 37°C). Next, acetonitrile was added in the ratio of 4:1 (v/v) and mixed for 5 min and subsequently centrifuged under 10,000 g for 5 min. The supernatant was collected for LC-MS analysis.
The interaction and pathway analysis of potential biomarkers was performed with Metacore version 4.3 (GeneGo Inc, St Joseph, MI). Network interaction analysis was performed on the basis of ‘shortest paths’ and the number of steps in a path were 4.
3 Results and discussion
3.1 Clinical chemistry and histopathological assessment
Various clinical parameters were measured in plasma in order to monitor the toxicity effects of DOX (Table 1). Actually, in the high dose group (DOX applied at 20 mg/kg), the concentration of ALT and AST showed significant increase in blood 24 h after administration; ALT, AST and CK increased 48 h after dosing, and finally all parameters changed significantly 96 h after dosing. However, these clinical parameters did not show a significantly change for the different time points at the middle (10 mg/kg) and low (5 mg/kg) dose, only for LDH a statistically significant change could be observed 96 h after dosing for the middle dose group.
With histopathological examination no significant changes in liver, kidney and heart in all groups were detected 24 h after administration of DOX. Forty-eight hours after administration, toxicity-related alterations including mild myocardial fibers swelling, liver cell hydropic degeneration and kidney tubules swelling were observed in the high dose group; in the middle-dose group there were no changes observed in hearts but mild swelling and hydropic degeneration was found in livers and kidneys, whereas no changes were observed in the low dose group. Ninety-six hours after administration, in the high dose and middle dose groups distinct organ-lesions were observed including interstitial oedema, subendocardial fibrosis and degeneration of the myocardium in hearts, centrilobular hepatocellular necrosis and vacuolation in livers and tubular necrosis, large hyaline casts in tubular lumen, cloudy swelling of the tubular degeneration, tubular dilatation glomerular congestion in the kidneys, whereas in the low dose group no changes were observed. The clinical chemistry and histopathology indicated the presence of dose and time dependent lesions to liver, heart and kidney due to the toxicity of DOX.
3.2 UPLC-MS
In this study a simple urine profiling method was aimed for rather than a prefractionation of urine and subsequent analysis of the various fractions. Therefore, for sample preparation the sample was only diluted with water, and analyzed with UPLC-MS using a linear gradient with 0.1% formic acid and acetonitrile as eluents. As more metabolites were detected in positive mode than with negative mode, positive mode was chosen in this study. Typical total ion chromatograms of rat urines (Fig. 1) showed information rich metabolite profiles; totally more than 2,700 chromatographic peaks could be detected in the various ion traces (after administration, DOX can be detected in urine sample with retention time of 14.7 min Fig. 1e). Metabolite profiles of post-dose urine obtained by UPLC-MS revealed significant metabolic alterations, especially 96 h after administration, a group of metabolites with retention times from 14 min to 18 min were significantly higher in the samples of the high and middle dose groups (Fig. 1a–d).
3.3 Multivariate data analysis
The data were analyzed using PCA to obtain a global view and to find the systemic metabolic changes with time and dose. Ninety-six hours after administration the metabolites with retention times from 14 min to 18 min were significantly higher in the high and middle dose samples (Fig. 1). First, the peaks of whole chromatogram (0.5–18 min) were used to construct a dataset for PCA analysis; it was found that the peaks eluting from 14 min to 18 min tend to have a dominant effect in the PCA analysis. To investigate this further, the peaks of the whole chromatogram were separated into two parts, the first part was from 0.5 min to 14 min, the second part was from 14 min to 18 min; and both datasets were subjected to PCA analysis. A better separation of groups according to time and dose was obtained by using the peaks acquired from 0.5 min to 14 min. Principal components 1 (PC1) and 2 (PC2) explained 40% and 20% of the total variance (Fig. 2). The urine samples that were collected prior to administration of DOX and the control samples were clustered together in the PCA scores plot, together with the low-dose group samples 24 h and 48 h after applying DOX. All remaining samples were separated from the control and prior dose samples. Compared with the clinical chemistry and histopathological assessment, the metabolomics approach provided a more sensitive method to detect effects of toxicity; for example, whereas there was no clear difference between the middle dose group at 24 and 48 h after administration for the clinical parameters compared to the control group, a difference between especially the middle dose group at 48 h and control group, but also to some extent between the middle dose group at 24 h and control group, was visible with the PCA score plot. For the middle dose group (and also high dose group) at 24 h, a difference was found with the PCA score plot compared to the control group, but not with the histopathological assessment. Fig. 2 showed that due to the toxicity of DOX, after dosing the metabolite profiles of dosed rats were different form that of undosed rats.
The toxicity effects induced by DOX are dose and time dependent (Singal and Iliskovic 1998; De Beer et al. 2001). Figure 2 shows that the high dose group shows a larger response on DOX, i.e. a larger deviation from the control group and prior administration group, than the middle and low dose groups for each time point after dosing. Two different trends of the samples with time in the PCA score plot could be observed: 24 h and 48 h after dosing a shift of all middle and high dose samples from the control and prior dosing samples towards the right upper quarter can be observed, which was highest for the high dose samples; 96 h after dosing, there was an additional shift superimposed towards the lower right quarter observed, which was again highest for the high dose and lowest for the low dose. Such a path of samples in the PCA score plot with time may be considered as a metabolic trajectory (Keun et al. 2004) and describes a systemic metabolic change (Keun et al. 2004; Holmes et al. 1992; Beckwith-hall et al. 1998).
There was no trend to recover from the dose effect, which might suggest that the induced lesion was irreversible. These results were confirmed by histopathology, where multiple organs necroses were observed at 96 h post dose, and to some extent at 48 h. The DOX induced toxicity might be interpreted to have a “time threshold”: for the low-dose group, from the PCA scores plot there was no separation after 24 and 48 h compared with the control group, only after 96 h, differences were observed. This result suggests that even a low dose of DOX still can cause toxicity effects, but the effect is expressed in the samples only after some time. The results of PCA analysis suggest that the toxicity effects of DOX are dose and time dependent, however, the different variations due to time and dose, and their possible interaction are difficult to separate using PCA.
To separate the dose and time effects and estimate whether the dose effect depends on the time (i.e. interaction effect between dose and time), ASCA was used. ASCA splits the original dataset into different datasets describing the main dose effect, main time effects and a dose-time interaction (Eq. 1) (Smilde et al. 2005; Jansen et al. 2005).
By separating the original dataset into several sub-datasets, submodels can be constructed to elucidate time, dose and time-dose interaction effects of administrating DOX to the rats. As for PCA, for ASCA only the peaks with retention times between 0.5 and 14 min were taken into account. It was found that the amount of variation related to time, dose and their interaction were 30%, 12% and 23%, respectively. The residual variation was 35%. This result indicates that time, dose and their interaction all contributed to the development of DOX induced toxicity and the variation related to time was dominant. Further, it would be helpful to investigate the ‘pure’ effect of dose, time or their interaction on the metabolite profiles. PC1 of the submodel for ‘dose’ (Fig. 3a) explained 86% of the variation on the factor of ‘dose’ and showed an increasing separation of low, middle to high-dose from the control group. Similar results were also found for the submodel for ‘time’, an increasing separation from 24 h before dosing was found according to 24 h, 48 h and 96 h after administration, in which PC1 explained 72% of the variation (Fig. 3b). The plots of PC1 over dose and time showed in both cases an increase of PC1 with dose or time, which indicated that the impact of DOX on the metabolite profiles increased with the dose and time (Fig. 3a, b). Investigation of the interaction model (the ‘dose × time’ model) (Fig. 3c) revealed the additional effects of DOX on top of the pure dose and time models. Clearly the time and dose effects were not independent of each other. The submodel of ‘dose × time’ accounted for 23% of original variation and PC1 explained 45% of the variation of the submodel. For the control, low-dose and middle-dose samples similar and rather constant patterns over time were observed; only the 96 h samples seemed to be slightly different from the previous time point samples. On the other hand, for the high-dose samples a different trend was observed. All high dose samples and especially the 96 h samples differed from the other samples. The significant separation of the high dose group from the others after 96 h indicates that the interaction effect mainly occurred 96 h after dose and mainly affects the high dose group (the separation of low and middle dose from control groups is small). Still, the samples were arranged along the PC1 axis according to high dose, middle dose, low dose and control group. For the 24 h and 48 h samples, separation of the high-dose group from the other groups was clear and larger than for the difference between high dose and other groups at −24 h (i.e. prior administration). These results show, unlike the linear model of dose and time effect, the effect of interaction of dose and time is most pronounced for the high dose group and the last time points (96 h after administration). In summary, differences between dose groups and time point found with the interaction ‘dose × time’ model were rather comparable to those which were obtained with the PCA model, but were more clearly visualized.
3.4 Biomarkers identification
With PCA data were visualized by plotting the PC scores where each point in the scores plot represents an individual sample and the PC loadings where each point represents one mass/retention time pair. Thus, the loadings plot gives an indication of the metabolites that most strongly influence the patterns in the scores plot. From the PCA loading plots, various metabolites could be identified as being responsible for the separation between control and different dose groups, and were therefore viewed as potential biomarkers. Two datasets were constructed in this study (see Sect. 2.6, i.e. 0.5–14 min and 0.5–18 min) and subjected to PCA analysis to screen possible biomarkers. The biomarkers screened from the two datasets complemented each other and 34 peaks were assigned as possible biomarkers. A Student’s t-test, was conducted whether significant changes were observed for these metabolites at various time points and various doses compared to the control group, and the metabolites for which the assigned identity could be confirmed with reference standards were listed in Table 2. The identification of these metabolites was achieved according to the following general procedure: (i) identification of the elemental composition by high resolution mass spectrometry (LC-TOF-MS or LC-LTQ-FTMS), (ii) database search for known metabolites for obtained elemental composition, (iii) elimination of certain candidates obtained from the library using the information obtained from MS/MS spectra and (iv) inclusion of retention time information. Actually, it has to be taken into account that the metabolite can be a conjugate, which is often not included in databases yet. For many elemental compositions a huge number of possible candidates can be found, and the proper candidate has to be identified, by rejecting possible, but not correct candidates. If the metabolite is not found in any database or all candidates were rejected, and if the metabolite is not a conjugate, fractionation and enrichment for NMR analysis is a good option. However, NMR analysis is often not sensitive enough for metabolites at low concentrations and if the amount of sample is limited (Rezzi et al. 2008). If the metabolite is a conjugate, fractionation and hydrolysis is an option, with subsequent LC-MS/MS analysis. In the final step, the assigned identity of the metabolite should be confirmed by the analysis of a reference compound if available; in this manner the identity of uric acid, tryptophan, phenylalanine, hippuric acid and cholic acid (see Table 2) was confirmed. We will discuss below two typical challenges we encountered during the identification procedure, i.e. the identification of the quasi-molecular ion in a complex mass spectrum and the identification of a biomarker being a conjugate or modified by biotransformation.
Though ESI is a relative soft ionization method, several dozen m/z ratios may be recorded for a single chromatographic peak. These ions may be different metabolites, in-source fragmentation products (Lafaye et al. 2003) or various adducts. As an example, peaks with m/z 355.2 and 373.2 at a retention time of 16.7 min increased significantly at 96 h after dosing (Fig. 1f). Many masses were observed in the mass spectrum including m/z 355.2, 373.2, 431.2, 817.4, 839.4, etc. Obtaining the proper quasi-molecular ion information is critical. Actually, a negative ion mode MS analysis in the same chromatography condition was helpful to provide complementary information: in negative ion mode, the mass spectrum of the peak with a retention time of 16.7 showed major ions at m/z 407.2 and 815.4, which may correspond to [M − H]− and [2M − H]−, respectively. These results suggest that the molecular mass of the biomarker should be 408.2. Actually no [M + H]+ ion was detected in positive mode, and the masses acquired in positive mode were assigned as follows: m/z 355.2, 373.2 were in source fragments, and m/z 431.2, 817.4 and 839.4 were [M + Na]+, [2M + H]+ and [2M + Na]+, respectively.
Often, biotransformed conjugates of metabolites are not present in databases. Such metabolites usually have typical neutral losses, such as the loss of 176 for glucuronic acid conjugate. In this study, a significant (with regards to occurrence in different groups and time points) metabolite with m/z 338.1 at a retention time of 7.16 min was an interesting biomarker with the neutral loss of 176 (Fig. 4), suggesting that the metabolite might be a glucuronic acid conjugate. As no possible glucuronic acid conjugate candidate could be found in the database for that nominal mass (Pubchem compound, http://www.ncbi.nlm.nih.gov/sites/entrez), the elemental composition of the molecular ion and characteristic fragments ions were determined by accurate mass determination with FTMS. The loss of 176.0320 corresponded indeed to the loss of glucuronic acid. The elemental composition obtained for the unconjugated metabolite was C9H7NO2. Next, a hydrolysis reaction with ß-glucuronidase of the appropriate fraction of the HPLC separation was carried out, and the obtained sample analyzed with HPLC-MS. A new peak with m/z 162.1 showed up eluting later than the conjugated metabolite, which actually is expected for a glucuronic acid conjugate. The number of candidate compounds with a formula of C9H7NO2 was found to be 179 with Pubmed. MS3 analysis of this glucuronic acid conjugate was performed and the loss of m/z 18 (162–144), m/z 46 (162–116), m/z 28 (144–116) and m/z 27 (116–89) indicated that this compound might contain a COOH group or one CO and one OH group. Next, by calculation of the degree of unsaturation (the double bond equivalent is 7), this compound might have a heterocyclic aromatic ring. In addition, the background information of the samples was taken into account, i.e. that this compound should be an endogenous metabolite from rat or a natural compound from food. By applying these criteria’s, potential candidates are reduced to two groups, one group were indole carboxylic acids including indole-2-carboxylic acid, indole-3-carboxylic acid, indole-5-carboxylic acid etc., another group were quinoline derivatives including quinoline-3,4-diol, 2,8-quinolinediol etc. For distinguishing these two groups, a negative MS/MS was helpful, as indole carboxylic acids have a typical –COOH group and will produce a significant loss of –COO (with m/z 44) peak (as confirmed by analysis of carboxylic acid standards). As the hydrolyzed biomarker did not show this typical mass peak, the potential candidates were reduced to quinoline derivatives. Finally, by comparing retention time and MS/MS fingerprint, this biotransformed metabolite was identified as 2,8-quinolinediol glucuronic acid conjugate (QGAC).
3.5 Biological interpretation
Table 2 lists the identified metabolites which endogenous levels have been altered as a consequence of DOX treatment. The metabolites might be considered as potential markers of DOX induced toxicity. For the biological connection between these metabolites and DOX toxicity a network was constructed using Metacore (Fig. 5). Except for 2,8-quinolinediol glucuronic acid (QGAC), all metabolites were present in the Metacore database (including DOX). It was found that all metabolites associated with the toxic effects of DOX could be linked to each other within four steps, i.e. a known biological connection was there between the metabolites associated with the toxic effects of DOX. However, in the network most compounds were linked to each other with four steps, except for phenylalanine and tryptophan (2 steps). In most cases the connectors were transport proteins with a broad specificity. In conclusion, the biological link in most cases was not very specific and cannot be seen as a strong biological connection. However, this finding is not completely surprising, as the biological interpretation is based on existing knowledge and is limited to the data of the database and the collection of the data; most data have been collected from different species and are thus not organism-specific (Bugrim et al. 2004). Furthermore, the biocomplexity of mammalian also makes direct connection of dynamic changed biomarkers difficult, highly complex animals can be considered ‘superorganisms’ with an internal ecosystem of diverse symbiotic microbiota and parasites that have interactive metabolic processes (Nicholson et al. 2004). Only with a systems toxicology approach, these indirect biomarkers can be identified. For example, QGAC is not in the database and shows a decreasing trend after dosing. 2,8-Quinolinediol has been reported as a metabolite in rat urine after being fed a diet containing corn but could not be isolated form corn; it was speculated that it must be formed from a precursor present in the corn (Inagami et al.1965). It was also reported that 2,8-quinolinediol could be transformed from quinoline by bacteria(Shukla 1986), which may suggest that 2,8-quinolinediol and QGAC could be derived from food and undergo biotransformation by gut microbiota or by mammalian tissues.
The biological function of the individual biomarkers may provide some clues for the understanding of DOX induced toxicity. From 24 h to 48 h after administration, drug induced toxicity disturbs the metabolism of the organism and causes a metabolic deviation from homeostasis. Biomarkers at this stage reflect the subtle dysfunction or adaptive response. Hippuric acid, QGAC and uric acid may be used to characterize the toxicity. Actually, in toxicity studies a decrease in urinary hippurate is often observed (Keun 2006). In rat hippurate is synthesized from benzoate and glycine by liver mitochondria and the pathway from benzoate to hippurate may reflect hepatic mitochondrial dysfunction induced by DOX (Gatley and Sherratt 1977; Berthiaume and Wallace 2007); also, gut microbial metabolism and diet can affect the levels of hippurate in urine (Nicholls et al. 2003; Wang et al. 2005). The possible reasons for the decrease may be due to different factors including the toxin having an antibiotic effect which may reduce urinary hippurate by suppressing gut microflora, or the toxin can compete with transport into the proximal tubular cells (Halligan et al. 1995). DOX lacked appreciable antibacterial activities (Adrien Bodet et al. 1985). Plausible reasons for the decreased hippurate could be the binding of DOX to the inner mitochondria membrane resulting in mitochondrial lesions and reduction of the synthesis of hippuric acid (Goormaghtigh et al. 1990), or the effects of DOX induced reactive oxygen species (ROS) on mitochondria(Berthiaume and Wallace 2007). Uric acid is another sensitive and early biomarker for monitoring the drug toxicity. Other researchers also found elevated uric acid concentrations in plasma when high doses of DOX were given to rat (Öz and İlhan 2006). Systemic administration of uric acid is known to increase serum antioxidant capacity and it can reduce oxidative stress (Waring et al. 2001; Waring et al. 2003). The increased level in biofluid could be useful to eliminate ROS and can reduce the toxicity of DOX. The QGAC has been reported as a peroxisome proliferator-activated receptor α-related biomarker (Zhen et al. 2007), the decrease of this compound may indicate the effect of ROS on the peroxisome (Dewald et al. 2005).
The characteristics of toxicity at 96 h point to an irreversible cell death and severe organ lesions. The increase of tryptophan and phenylalanine in urine has been found in d-serine-induced nephrotoxicity and were considered as nephrotoxicity biomarkers (Williams et al. 2005) and indicated the dysfunction of kidney. Cholic acid is one of bile acids derived from cholesterol, primarily in the liver, through three main pathways involving different enzymes (Björkhem and Eggertsen 2001) and can be used as a marker of liver dysfunction. Administration of compounds known to specifically cause damages to the biliary system in the liver can result in an increase of bile acid in urine (Beckwith-Hall et al. 1998). In this study, at 96 h post dose, cholic acid increased significant for high and middle dose group. Even for low dose group, three animals showed a high level of cholic acid. This result indicates that DOX can induce liver damage.
4 Conclusions
An UPLC-MS based metabolomics approach was used to investigate the toxicity induced by DOX in a rat model. Significant dose and time dependent changes were found in the UPLC-MS urine metabolite profiles. With the PCA model, two major systemic metabolic changes with time, or metabolic trajectories, could be found for the development of the toxicity induced by DOX. ASCA allowed separating the original dataset into submodels to interpret the systemic effects derived from dose, time and their interactions. Some possible exploratory early toxicity biomarkers due to DOX were found. In conclusion, the proposed systems toxicity approach based on metabolomic profiling was able to reveal novel information in the toxicity of DOX. In addition, detection of systemic toxic effects by metabolomics appeared to be more sensitive, i.e. detection was possible at a more early stage, compared with clinical chemistry and histopathological assessment.
References
Adrien Bodet, C., III, Jorgensen, J. H., & Drutz, D. J. (1985). Antibacterial activities of antineoplastic agents. Antimicrobial Agents and Chemotherapy, 28, 437–439.
Bagchi, D., Bagchi, M., Hassoun, E. A., Kelly, J., & Stohs, S. J. (1995). Adriamycin-induced hepatic and myocardial lipid peroxidation and DNA damage, and enhanced excretion of urinary lipid metabolites in rats. Toxicology, 95, 1–9.
Beckwith-Hall, B. M., Nicholson, J. K., Nicholls, A. W., et al. (1998). Nuclear magnetic resonance spectroscopic and principal components analysis investigations into biochemical effects of three model hepatotoxins. Chemical Research in Toxicology, 11, 260–272.
Berthiaume, J. M., & Wallace, K. B. (2007). Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biology and Toxicology, 23, 15–25.
Björkhem, I., & Eggertsen, G. (2001). Genes involved in initial steps of bile acid synthesis. Current Opinion in Lipidology, 12, 97–103.
Bugrim, A., Nikolskaya, T., & Nikolsky, Y. (2004). Early prediction of drug metabolism and toxicity: Systems biology approach and modeling. Drug Discovery Today, 9, 127–135.
De Beer, E. L., Bottone, A. E., & Voest, E. E. (2001). Doxorubicin and mechanical performance of cardiac trabeculae after acute and chronic treatment: A review. European Journal of Pharmacology, 415, 1–11.
Dewald, O., Sharma, S., Adrogue, J., et al. (2005). Downregulation of peroxisome proliferator-activated receptor-alpha gene expression in a mouse model of ischemic cardiomyopathy is dependent on reactive oxygen species and prevents lipotoxicity. Circulation, 112, 407–415.
Gatley, S. J., & Sherratt, H. S. A. (1977). The synthesis of hippurate from benzoate and glycine by rat liver mitochondria. Submitochondrial localization and kinetics. The Biochemical Journal, 166, 39–47.
Goormaghtigh, E., Huart, P., Praet, M., Brasseur, R., & Ruysschaert, J. M. (1990). Structure of the adriamycin–cardiolipin complex. Role in mitochondrial toxicity. Biophysical Chemistry, 35, 247–257.
Halligan, S., Byard, S. J., Spencer, A. J., Gray, T. J. B., Harpur, E. S., & Bonner, F. W. (1995). A study of the nephrotoxicity of three cephalosporins in rabbits using 1H NMR spectroscopy. Toxicology Letters, 81, 15–21.
Holmes, E., Bonner, F. W., Sweatman, B. C., et al. (1992). Nuclear magnetic resonance spectroscopy and pattern recognition analysis of the biochemical processes associated with the progression of and recovery from nephrotoxic lesions in the rat induced by mercury(II) chloride and 2-bromoethanamine. Molecular Pharmacology, 42, 922–930.
Inagami, K., Kaihara, M., & Price, J. M. (1965). The identification of 2,8-quinolinediol in the urine of rats fed a diet containing corn. The Journal of Biological Chemistry, 240, 3682–3684.
Jansen, J. J., Hoefsloot, H. C. J., van der Greef, J., Timmerman, M. E., Westerhuis, J. A., & Smilde, A. K. (2005). ASCA: analysis of multivariate data obtained from an experimental design. Journal of Chemometrics, 19, 469–481.
Kalender, Y., Yel, M., & Kalender, S. (2005). Doxorubicin hepatotoxicity and hepatic free radical metabolism in rats. The effects of vitamin E and catechin. Toxicology, 209, 39–45.
Kamphorst, J. J., van der Heijden, R., DeGroot, J., et al. (2007). Profiling of endogenous peptides in human synovial fluid by NanoLC-MS: Method validation and peptide identification. Journal of Proteome Research, 6, 4388–4396.
Keun, H. C. (2006). Metabonomic modeling of drug toxicity. Pharmacology & Therapeutics, 109, 92–106.
Keun, H. C., Ebbels, T. M. D., Bollard, M. E., et al. (2004). Geometric trajectory analysis of metabolic responses to toxicity can define treatment specific profiles. Chemical Research in Toxicology, 17, 579–587.
Lafaye, A., Junot, C., Ramounet-Le Gall, B., Fritsch, P., Tabet, J. C., & Ezan, E. (2003). Metabolite profiling in rat urine by liquid chromatography/electrospray ion trap mass spectrometry. Application to the study of heavy metal toxicity. Rapid Communications in Mass Spectrometry, 17, 2541–2549.
Lenz, E. M., Bright, J., Knight, R., Wilson, I. D., & Major, H. (2004). Cyclosporin A-induced changes in endogenous metabolites in rat urine: a metabonomic investigation using high field 1H NMR spectroscopy, HPLC-TOF/MS and chemometrics. Journal of Pharmaceutical and Biomedical Analysis, 35, 599–608.
Lindon, J. C., Nicholson, J. K., Holmes, E., et al. (2003). Contemporary issues in toxicology the role of metabonomics in toxicology and its evaluation by the COMET project. Toxicology and Applied Pharmacology, 187, 137–146.
Meliksetian, M. B., Daviyan, T. K., Chirkowa, I. V., Alexanyan, Y. U. T., & Ignatova, T. N. (1997). Alterations in immunoglobulin synthesis during adriamycin-induced apoptosis in mouse hybridoma cells. Cell Biology International, 21, 69–74.
Nicholls, A. W., Mortishire-Smith, R. J., & Nicholson, J. K. (2003). NMR spectroscopic-based metabonomic studies of urinary metabolite variation in acclimatizing germ-free rats. Chemical Research in Toxicology, 16, 1395–1404.
Nicholson, J. K., Holmes, E., Lindon, J. C., & Wilson, I. D. (2004). The challenges of modeling mammalian biocomplexity. Nature Biotechnology, 22, 1268–1274.
Nicholson, J. K., Lindon, J. C., & Holmes, E. (1999). ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica, 29, 1181–1189.
Oliveira, P. J., Bjork, J. A., Santos, M. S., et al. (2004). Carvedilol-mediated antioxidant protection against doxorubicin-induced cardiac mitochondrial toxicity. Toxicology and Applied Pharmacology, 200, 159–168.
Öz, E., & İlhan, M. N. (2006). Effects of melatonin in reducing the toxic effects of doxorubicin. Molecular and Cellular Biochemistry, 286, 11–15.
Rezzi, S., Vera, F. A., Martin, F. P., Wang, S., Lawler, D., & Kochhar, S. (2008). Automated SPE-RP-HPLC fractionation of biofluids combined to off-line NMR spectroscopy for biomarker identification in metabonomics. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences, 871, 271–278.
Saad, S. Y., Najjar, T. A., & Al Rikabi, A. C. (2001). The preventive role of deferoxamine against acute doxorubicin-induced cardiac, renal and hepatic toxicity in rats. Pharmacological Research, 43, 211–218.
Shukla, O. P. (1986). Microbial transformation of quinoline by a Pseudomonas sp. Applied and Environmental Microbiology, 51, 1332–1342.
Singal, P. K., & Iliskovic, N. (1998). Doxorubicin-induced cardiomyopathy. The New England Journal of Medicine, 339, 900–905.
Smilde, A. K., Jansen, J. J., Hoefsloot, H. C. J., Lamers, R. A. N., van der Greef, J., & Timmerman, M. E. (2005). ANOVA-simultaneous component analysis (ASCA): A new tool for analyzing designed metabolomics data. Bioinformatics, 21, 3043–3048.
Swift, L. P., Rephaeli, A., Nudelman, A., Phillips, D. R., & Cutts, S. M. (2006). Doxorubicin-DNA adducts induce a non-topoisomerase II-mediated form of cell death. Cancer Research, 9, 4863–4871.
Theodoridis, G., Gika, H. G., & Wilson, I. D. (2008). LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trends in Analytical Chemistry, 27, 251–260.
Van der Greef, J., Martin, S., Juhasz, P., et al. (2007). The art and practice of systems biology in medicine: mapping patterns of relationships. Journal of Proteome Research, 6, 1540–1559.
Van der Greef, J., Stroobant, P., & van der Heijden, R. (2004). The role of analytical sciences in medical systems biology. Current Opinion in Chemical Biology, 8, 559–565.
Wang, Y., Tang, H., Nicholson, J. K., Hylands, P. J., Sampson, J., & Holmes, E. (2005). A metabonomic strategy for the detection of the metabolic effects of chamomile (Matricaria recutita L.) ingestion. Journal of Agricultural and Food Chemistry, 53(19), 1–196.
Wapstra, F. H., van Goor, H., de Jong, P. E., Navis, G., & de Zeeuw, D. (1999). Dose of doxorubicin determines severity of renal damage and responsiveness to ACE-inhibition in experimental nephrosis. Journal of Pharmacological and Toxicological Methods, 41, 69–73.
Waring, W. S., Convery, A., Mishra, V., Shenkin, A., Webb, D. J., & Maxwell, S. R. J. (2003). Uric acid reduces exercise-induced oxidative stress in healthy adults. Clinical Science, 105, 425–430.
Waring, W. S., Webb, D. J., & Maxwell, S. R. (2001). Systemic uric acid administration increases serum antioxidant capacity in healthy volunteers. Journal of Cardiovascular Pharmacology, 38, 365–371.
Waters, N. J., Waterfield, C. J., Farrant, R. D., Holmes, E., & Nicholson, J. K. (2005). Metabonomic deconvolution of embedded toxicity: application to thioacetamide hepato- and nephrotoxicity. Chemical Research in Toxicology, 18, 639–654.
Williams, R. E., Major, H., Lock, E. A., Lenz, E. M., & Wilson, I. D. (2005). D-Serine-induced nephrotoxicity: A HPLC-TOF/MS-based metabonomics approach. Toxicology, 207, 179–190.
Yang, J., Zhao, X., Liu, X., et al. (2006). High performance liquid chromatography-mass spectrometry for metabonomics: Potential biomarkers for acute deterioration of liver function in chronic hepatitis B. Journal of Proteome Research, 5, 554–561.
Zhen, Y., Krausz, K. W., Chen, C., Idle, J. R., & Gonzalez, F. J. (2007). Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor alpha expression and activation. Molecular Endocrinology, 21, 2136–2151.
Acknowledgements
The support of JW by a Royal Netherlands Academy of Science-Chinese Academy of Sciences Joint Research Project Grant (05CDP010) is very much acknowledged. This study is also supported by the China International Science and Technology Cooperation Program (2007DFA31060) and National Key Technologies R&D Program (2006038079037) and National Key Project on Drug Development from the Ministry of Science and Technology of China. Dr. Suzan Wopereis (TNO Quality of Life, Zeist, The Netherlands) is acknowledged for discussion of the biological interpretation of the results and supplying the Metacore analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Wang, J., Reijmers, T., Chen, L. et al. Systems toxicology study of doxorubicin on rats using ultra performance liquid chromatography coupled with mass spectrometry based metabolomics. Metabolomics 5, 407–418 (2009). https://doi.org/10.1007/s11306-009-0165-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-009-0165-3