Abstract
In the last few years, many efforts have been made to search for potent and selective human A3 adenosine antagonists. In particular, one of the most promising human A3 adenosine receptor antagonists is represented by the pyrazolo-triazolo-pyrimidine family. This class of compounds has been strongly investigated from the point of view of structure-activity relationships. In particular, it has been observed that fundamental requisites for having both potency and selectivity at the human A3 adenosine receptors are the presence of a small substituent at the N8 position and an unsubstitued phenyl carbamoyl moiety at the N5 position. In this study, we report the role of the N5-bond type on the affinity and selectivity at the four adenosine receptor subtypes. The observed structure-activity relationships of this class of antagonists are also exhaustively rationalized using the recently published ligand-based homology modeling approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adenosine is an ubiquitous modulator, which exerts its functions through interaction with four G-protein-coupled receptors classified as A1, A2A, A2B and A3 [1]. In recent decades, intensive and successful efforts have been made by medicinal chemists to discover potent and selective ligands (both agonists and antagonists) for almost all adenosine receptor subtypes [2, 3]. Only the A2B subtype is still without a very potent and selective agonist ligand [4–6]. At the same time, great progress has been made in the field of human A3 adenosine receptor antagonists. This receptor seems to be associated with cerebroprotection [7, 8] and cardioprotection [9] and effects on the immune and inflammatory systems [10, 11]. The A3 adenosine receptor subtype, recently cloned from different species (e.g., rat, human, dog, sheep) [12, 13], is coupled to the modulation of at least two second-messenger systems: inhibition of adenylate cyclase and stimulation of phospholipase C and D [12, 13]. In humans, A3 receptors have been found in several organs, such as lung, liver, kidney, heart, and with a lower density, in the brain [9, 12, 13]. This receptor subtype is under examination in relation to its potential therapeutic applications. In particular, antagonists for A3 receptors seem to be useful for the treatment of inflammation or glaucoma [14, 15].
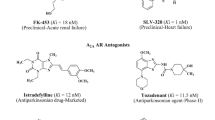
In the last 5 years, many efforts have been made in the context of searching for potent and selective human A3 adenosine antagonists. In this field, several different classes of compounds have been proposed, possessing good affinity (nM range) and with a broad range of selectivity [16].
In particular one of the most attractive human A3 adenosine receptor antagonists is represented by the pyrazolo-triazolo-pyrimidine family, reported by our group [17–23]. In Fig. 1, the most interesting members of this class of compounds (1–3), which display high affinity and selectivity for this receptor subtype, are depicted [17–23].
This class of compounds has been strongly investigated from the point of view of structure-activity relationships. In particular, it has been observed that fundamental requisites for having both potency and selectivity at the human A3 adenosine receptors are a small substituent at the N8 position and an unsubstituted phenyl carbamoyl moiety at the N5 position [17, 21]. In contrast, when the phenyl carbamoyl moiety (2) was replaced with a phenylacetyl group (3), a significant decrease in affinity at the hA3 adenosine receptors with a simultaneous retention or a slight increase in affinity at the hA2B adenosine receptor subtype was observed [24].
These data seem to suggest that bond type at the N5 position plays a fundamental role in the receptor recognition. For this reason, we decided to further investigate the effect of the bond nature at this position on affinity and selectivity, synthesizing two new derivatives bearing the benzoyl- (4) and benzensulfonamido (5) moieties at the N5 position.
Results and discussion
Newly synthesized (4, 5) and reference compounds (2, 3) were tested at the human A1, A2A and A3 receptors expressed in CHO cells; [3H]CCPA (A1) and [3H]NECA (A2A, A3) were used as radioligands in binding assays [25]. Radioligand binding at A2B adenosine receptors is problematic as no high-affinity ligand is readily available for this subtype. Therefore, inhibition of NECA-stimulated adenylyl cyclase activity was determined as a measurement of affinity of compounds (Table 1).
A careful examination of the data clearly revealed significant differences in the binding profile of the reference compounds (2, 3) compared with the literature data (see Fig. 1). In fact, in these binding studies, compound 2 was found to be ninefold less potent at the hA3 receptors with a consequent reduction in selectivity versus the other receptor subtypes, in particular versus hA2A (hA2A/hA3 = 15). In contrast, derivative 3 retained good affinity at the hA3 adenosine receptor and a quite good range of selectivity versus the other receptors in comparison with literature data. Also if these discrepancies in binding profile could be attributed to a different biological method utilized (e.g., radioligand), the data seem to indicate that the phenylacetyl moiety retains or enhances both potency and selectivity compared with the well-known phenylcarbamoyl chain. A significantly different binding profile was instead observed when the chain at the N5 position was alternatively modified. Introduction of a shorter amide (4) with respect phenylacetyl chain (3) led to a loss of potency at all four adenosine receptor subtypes, but a quite good retention of selectivity was observed. In contrast, the presence of a benzensulfonamido moiety at the N5 position induced a substantial loss of both potency and selectivity at the four adenosine receptor subtypes.
Molecular modeling studies were performed on the pyrazolo-triazolo-pyrimidine derivatives 2–5 in order to identify the hypothetical binding motif of these N5 analogues and to rationalize their structure-activity relationship.
Following our previously reported modeling studies [17–23], we built up a refined model of human A3 receptor by using a rhodopsin-based homology modeling (RBHM) approach [26–29]. Moreover, our recently described ligand-based homology modeling methodology (LBHM) has been used to simulate the conformational changes induced by ligand binding (methodological details are summarized in the Experimental Section) [29, 30].
Using this methodology, we found an “expanded” conformational model of the human A3 receptor reverse agonist-like state, in which both shape and chemical complementarities have been specifically optimized around each ligand. Considering these new N5 analogues, the molecular volume of transmembrane (TM) binding cavity has been changed from 660 (A3 model obtained by the conventional rhodopsin-based homology modeling) to 840 Å3 (expanded A3 model obtained by ligand-based homology modeling) without altering the conventional rhodopsin-like receptor topology. The binding cavity reorganization induced by ligand binding is due to the conformational change in several amino acid side chains, such as Leu90 (3.32), Leu91 (3.33), Thr94 (3.36), His95 (3.37), Ile98 (3.40), Gln167 (EL2), Phe168 (EL2), Phe182 (5.43), Ile186 (5.47), Leu190 (5.51), Phe239 (6.44), Trp243 (6.48), Leu244 (6.49), Leu264 (7.35), and Ile268 (7.39).
Interestingly, none of the new pyrazoloquinoline antagonists found an energetically stable docking pose in the conventional RBHM-driven A3 model. This is mainly due to the unfavorable topological complementarity among these antagonists and corresponding RBHM-driven TM binding cavity. In particular, highly destabilizing van der Waals interactions (steric conflicts) seem to be the reason for absent topological complementarities. These steric conflicts are drastically reduced or completely eliminated after the application of the LBHM approach.
Molecular docking studies were carried out for the pyrazolo-triazolo-pyrimidine antagonists 2–4, using the “expanded” conformational state of the receptor. As shown in Fig. 2, we found a similar binding motif indicating that a common receptor-driven pharmacophore model can be depicted. This finding is in agreement with our previously reported studies [17–23].
Hypothetical binding motif of the newly synthesized pyrazolo-triazolo-pyrimidine antagonists 2–4. The most energetically favorable docked conformation of each derivative is viewed from the membrane side facing TM helices 4 and 5. To clarify the TM cavity, the view of TM4 was omitted. Side chains of some amino acids important for ligand recognition are highlighted. Hydrogen atoms are not displayed
Indeed, ligand recognition occurs in the upper region of the TM bundle, and the pyrazolo-triazolo-pyrimidine moiety is surrounded by TMs 3, 5, 6, 7 with the substituent in the N5 position oriented toward the intracellular environment. As shown in Fig. 2, the furan ring at the 2-position is close to TMs 3 and 7. Interestingly, an important hydrogen bonding network can be observed in all energetically stable docked conformations of pyrazolo-triazolo-pyrimidine antagonists. In particular His95 (3.37) and Ser247 (6.52) are able to interact through hydrogen bonding with the N5-carbonyl oxygen of compounds 2–4 (2C=O·His95 ca. 3.0 Å; 3C=O·Ser247 ca. 2.8 Å; 4C=O·His95 ca. 2.9 Å.)
These polar amino acids seem to be critical for the recognition of all antagonist structures and for receptor selectivity. In particular, Ser247 (6.52) of human A3 receptor subtype is not present in the corresponding position of A1 and A2 receptors, where the residue is replaced by a histidine (His251 in human A1, His250 in human A2A and His251 in human A2B). Histidine side chain is bulkier than serine, and possibly for this reason, large substituents at the N5 position of pyrazolo-triazolo-pyrimidine framework are not well tolerated by A1 and A2 receptor subtypes. In contrast, the hydroxyl group of Ser247 (6.52) of human A3 receptor is appropriately positioned to form a hydrogen-bonding interaction with the carbonyl oxygen of the N5-amide/ureide group of compounds 2–4. These observations support the importance of an N5-acyl/carbamoyl group in modulating receptor selectivity. The hydrophobic environment of the five nonpolar amino acids Ile98 (3.40), Ile186 (5.47), Leu190 (5.51), Phe239 (6.44), and Leu244 (6.49) can comfortably accommodate the phenyl ring of all N5-acyl/carbamoyl derivatives.
In contrast, the introduction of the N5-sulfonamido moiety, as present in derivative 5, drastically reduces the affinity at the human A3 receptor. Interestingly, in this specific case molecular docking is not able to find an antagonist pose comparable to those described for the other N5-acyl/carbamoyl derivatives. As shown in Fig. 3, the rigid tetrahedral configuration associated with the N5-sulfonamido moiety avoids the sampling of energetically favorable antagonist poses in which the phenyl ring is linked to the N5 position in the hydrophobic pocket delimited by Ile98 (3.40), Ile186 (5.47), Leu190 (5.51), Phe239 (6.44), and Leu244 (6.49).
Hypothetical binding motif of the newly synthesized N5-sulfonamido pyrazolo-triazolo-pyrimidine antagonist 5. The most energetically favorable docked conformation of each derivative is viewed from the membrane side facing TM helices 4 and 5. To clarify the TM cavity, the view of TM4 was omitted. Hydrogen atoms are not displayed
The most stable docking pose of 5 presents the N5-sulfonamido moiety close to TM3 and TM7, and the phenyl ring linked to N5 position is surrounded by a hydrophobic pocket delimited by Leu90 (3.32) and Ile268 (7.39). This antagonist pose is energetically less stable (ca. 15 kcal/mol) with respect to those found for derivatives 2–4, due to the absence of the stabilizing interactions among the polar residues Thr94 (3.36), His95 (3.37), and Ser247 (6.52) and the N5-sulfonamido moiety. Structure superimposition of compounds 4 and 5 is shown in Fig. 4.
This severe steric constriction might explain the drastic reduction in affinity of derivative 5 at the human A3 receptor.
Conclusions
The present study has led to potent and selective human A3 adenosine receptor antagonists belonging to the class of the pyrazolo-triazolo-pyrimidine. In this study, we clarified the role of the N5-bond type on the affinity and selectivity at the four adenosine receptor subtypes. In particular, we confirmed the critical function played by the N5-acyl/carbamoyl moiety in receptor recognition. The observed structure-activity relationship was also rationalized by the recently published ligand-based homology modeling approach, indicating that chemical and topological properties of the different ligands might induce important conformational changes in the antagonist-driven binding site of the human A3 adenosine receptor.
Experimental section
Chemistry
General
Reactions were routinely monitored by thin-layer chromatography (TLC) ion silica gel (precoated F254 Merck plates) and products visualized with iodine or potassium permanganate solution. Infrared spectra (IR) were measured on a Perkin Elmer 257 instrument. 1H NMR was determined in CDCl3 or DMSO-d6 solutions with a Bruker AC 200 spectrometer; peak positions are given in parts per million (6) downfield from tetramethylsilane as internal standard, and J values are given in Hz. Light petroleum ether refers to the fractions boiling at 40–60°C. Melting points were determined on a Buchi-Tottoli instrument and are uncorrected. Chromatographies were performed using Merck 60–200 mesh silica gel. All products reported showed IR and 1H NMR spectra in agreement with the assigned structures. Organic solutions were dried over anhydrous magnesium sulphate. Elemental analyses were performed by the microanalytical laboratory of Dipartimento di Chimica, University of Trieste.
General procedures for the preparation of 5-(benzoyl)amino-8-methyl-2-(2-furyl)-pyrazolo[4,3 ]-1,2,4-triazolo[1,5-c]pyrimidine (4) and 5-benzensulfonamido-8-methyl-2-(2-furyl)-pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidine (5)
Amino compound (6) (10 mmol) was dissolved in freshly distilled dioxane (15 mL), and benzoylchloride or benzesulfonyl chloride (13 mmol) and pyridine (20 mmol) were added. The mixture was refluxed under argon for 16 h. Then the solvent was removed under reduced pressure and the residue was dissolved in EtOAc (30 ml) and washed twice with water (15 mL). The organic phase was dried on Na2SO4 and concentrated under reduced pressure. The residue was purified by flash chromatography (EtOAc-light petroleum 8:2) to afford the desired compounds (4, 5) as solids.
5-(Benzoyl)amino-8-methyl-2-(2-furyl)-pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidine (4)
Yield 78%, pale yellow solid, mp (EtOAc-light petroleum) 240°C; IR (KBr): 3,425-2,965, 1,675, 1,645, 1,620, 1,545, 1,450 cm−1; 1 H NMR (DMSO d6) δ: 4.42 (s, 3H); 6.65 (dd, 1H, J = 2, J = 4); 7.24 (d, 1H, J = 4); 7.41–7.62 (m, 4H); 7.85–7.98 (m, 2H); 8.14 (s, 1H); 9.87 (bs, 1H). Anal. for C18H13N7O2 (MW 359.34). Calculated: C 50.16; H 3.69; N 27.29. Found: C 50.01; H 3.61; N 27.35.
5-Benzensulfonamido-8-methyl-2-(2-furyl)-pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidine (5)
Yield 62%, white solid, mp (EtOAc-light petroleum) > 300°C; IR (KBr): 3,375-2,985, 1,640, 1,600, 1,540, 1,355, 1,150 cm−1; 1H NMR (DMSO d6) δ: 4.39 (s, 3H); 6.68 (dd, 1H, J = 2, J = 4); 7.22 (d, 1H, J = 4); 7.38–7.59 (m, 4H); 7.88-8.02 (m, 2H); 8.11 (s, 1H); 11.05 (bs, 1H). Anal. for C17H13N7O3S (MW 395.40). Calculated: C 51.64; H 3.31; N 24.80; S 8.11. Found: C 51.87; H 3.35; N 24.97; S 8.07.
Biology
All pharmacological methods followed the procedures as described earlier [25]. In brief, membranes for radioligand binding were prepared from CHO cells stably transfected with human adenosine receptor subtypes in a two-step procedure. In a first low-speed step (1,000 × g), cell fragments and nuclei were removed. The crude membrane fraction was sedimented from the supernatant at 100,000 × g. The membrane pellet was resuspended in the buffer used for the respective binding experiments, frozen in liquid nitrogen and stored at −80°C. For the measurement of adenylyl cyclase activity only one high-speed centrifugation of the homogenate was used. The resulting crude membrane pellet was resuspended in 50 mM Tris/HCl, pH 7.4 and immediately used for the cyclase assay.
For radioligand binding at A1 adenosine receptors, 1 nM [3H]CCPA was used, whereas 30 and 10 nM [3H]NECA were used for A2A and A3 receptors, respectively. Nonspecific binding of [3H]CCPA was determined in the presence of 1 mM theophylline, and in the case of [3H]NECA, 100 pM R-PIA was used. Ki values from competition experiments were calculated with the program SCTFIT [31].
Radioligand binding at A2B adenosine receptors is problematic as no high-affinity ligand is available for this subtype. Therefore, inhibition of NECA-stimulated adenylyl cyclase activity was determined as a measurement of affinity of compounds. IC50 values from these experiments were converted to Ki values with the Cheng and Prusoff equation [32].
Computational methodologies
All modeling studies were carried out on a 10 CPU (PIV-3.0GHZ and AMD64) Linux cluster running under openMosix architecture [33].
Homology modeling, energy calculation, and docking studies were performed using the Molecular Operating Environment (MOE, version 2006.08) suite [34].
All docked structures were fully optimized without geometry constraints using RHF/AM1 semiempirical calculations. Vibrational frequency analysis was used to characterize the minima stationary points (zero imaginary frequencies). The software package MOPAC (ver. 7) [35], implemented in MOE suite, was utilized for all quantum mechanical calculations.
Homology model of the human A3 AR
Based on the assumption that GPCRs share similar TM boundaries and overall topology, a homology model of the hA3 receptor was constructed. First, the amino acid sequences of TM helices of the A3 receptor were aligned with those of bovine rhodopsin, guided by the highly conserved amino acid residues, including the DRY motif (D3.49, R3.50, and Y3.51) and three proline residues (P4.60, P6.50, and P7.50) in the TM segments of GPCRs. The same boundaries were applied for the TM helices of the A3 receptor as they were identified from the X-ray crystal structure for the corresponding sequences of bovine rhodopsin [36], the CR coordinates of which were used to construct the seven TM helices for the hA3 receptor.
The loop domains of the hA3 receptor were constructed by the loop search method implemented in MOE. In particular, loops are modeled first in random order. For each loop, a contact energy function analyzes the list of candidates collected in the segment searching stage, taking into account all atoms already modeled and any atoms specified by the user as belonging to the model environment. These energies are then used to make a Boltzmann-weighted choice from the candidates, the coordinates of which are then copied to the model. Any missing side-chain atoms are modeled using the same procedure. Side chains belonging to residues whose backbone coordinates were copied from a template are modeled first, followed by side chains of modeled loops. Outgaps and their side chains are modeled last.
Special caution has to be given to the second extracellular (EL2) loop, which has been described in bovine rhodopsin as folding back over transmembrane helices [36] and, therefore, limiting the size of the active site. Hence, amino acids of this loop could be involved in direct interactions with the ligands. A driving force for this peculiar fold of the EL2 loop might be the presence of a disulfide bridge between cysteines in TM3 and EL2. Since this covalent link is conserved in all receptors modeled in the current study, the EL2 loop was modeled using a rhodopsin-like constrained geometry around the EL2-TM3 disulfide bridge. After the heavy atoms were modeled, all hydrogen atoms were added, and the protein coordinates were then minimized with MOE using the AMBER94 force field [37]. The minimizations were carried out by the 1,000 steps of steepest descent followed by conjugate gradient minimization until the rms gradient of the potential energy was less than 0.1 kcal mol−1 Å−1. Protein stereochemistry evaluation was performed by several tools (Ramachandran and Chi plots measure phi/psi and chi1/chi2 angles, clash contacts reports) implemented in MOE suite [34].
Ligand-based homology modeling
We have recently revisited the rhodopsin-based model of the human A3 receptor in its resting state (antagonist-like state), taking into account a novel strategy to simulate the possible receptor reorganization induced by the antagonist-binding [28]. We called this new strategy ligand-based homology modeling. Briefly, ligand-based homology modeling technique is an evolution of a conventional homology modeling algorithm based on a Boltzmann-weighted randomized modeling procedure adapted from Levitt [38] with specialized logic for the proper handling of insertions and deletions so that any selected atoms will be included in the energy tests and minimization stages of the modeling procedure. Ligand-based option is very useful when one wishes to build a homology model in the presence of a ligand docked to the primary template, or other proteins known to be complexed with the sequence to be modeled [34]. In this specific case, both model building and refinement take into account the presence of the ligand in terms of specific steric and chemical features. In order to generate an initial ensemble of ligand poses, a conventional docking procedure (see next section for details) with reduced van der Waals radii (equal to 75%) and an increased Coulomb-vdW cutoff (cutoff on 10 Å; cutoff on 12 Å) was performed. For each pose, an homology model is then generated to accommodate the ligand by reorienting nearby side chains. These residues and the ligand are then locally minimized. Finally, each ligand is re-docked into its corresponding low energy protein structures and the resulting complexes are ranked according to MOEScore [34].
Different quantitative measures of molecular volume of the receptor binding cavities have been carried out by using MOE suite [34]. Prediction of antagonist-receptor complex stability (in terms of corresponding pKi value) and the quantitative analysis for non-bonded intermolecular interactions (H-bonds, transition metal, water bridges, hydrophobic) were calculated and visualized using several tools implemented into MOE suite [34].
Molecular docking of the hA3 AR antagonists
All antagonist structures were docked into the hypothetical TM binding site by using the MOE-dock tool, part of the MOE suite. Searching is conducted within a user-specified 3D docking box (the standard protocol selects all atoms inside 12 Å from the center of mass of the binding cavity), using the Tabu Search [39] protocol (standard parameters are 1,000 steps/run, 10 attempts/step, and 10 Tabu list length), and the MMFF94 force field [40]. MOE-dock performs a user-specified number of independent docking runs (50 in our specific case) and writes the resulting conformations and their energies in a molecular database file. The resulting docked complexes were subjected to MMFF94 energy minimization until the rms of conjugate gradient was <0.1 kcal mol−1 Å−1. Charges for the ligands were imported from the MOPAC output files. To better refine all antagonist-receptor complexes, a rotamer exploration of all side chains involved in the antagonist-binding was carried out. Rotamer exploration methodology was implemented in MOE suite [34].
Abbreviations
- CHO cells:
-
chinese hamster ovary cells
- EtOAc:
-
ethyl acetate
- TLC:
-
thin layer chromatography
- NMR:
-
nuclear magnetic resonance
- IR:
-
infrared spectra
- CDC13 :
-
deuterated chloroform
- Na2SO4 :
-
sodium sulphate
- mp:
-
melting point
- KBr:
-
potassium bromide
- DMSO-d6 :
-
deuterated dimethylsulfoxide
- CCPA:
-
2-Chloro-N6-cyclopentyladenosine
- NECA:
-
5′-(N-Ethylcarbamoyl)adenosine
- R-PIA:
-
R(-)-N6-Cyclopentyladenosine
References
Fredholm BB, Ijzerman AP, Jacobson KA, Klotz KN, Linden J (2001) International Union of Pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol Rev 53:527–552
Baraldi PG, Cacciari B, Romagnoli R, Merighi S, Varani K, Borea PA, Spalluto G (2000) A3 Adenosine receptor ligands; history and perspectives. Med Res Rev 20:103–128
Jacobson KA, Knutsen LJS (2001) P1 and P2 purine and pyrimidine receptor ligands. In: Abbracchio MP, Williams M (eds) Handbook of experimental pharmacology, vol. 151. Purinergic and pyrimidinergic signalling I. Springer, Berlin, pp 129–175
Volpini R, Costanzi S, Vittori S, Cristalli G, Klotz KN (2003) Medicinal chemistry and pharmacology of A2b adenosine receptors. Current Topics Med Chem 3:427–443
Feoktistov I, Biaggioni I (1998) Pharmacological characterization of adenosine A2B receptors. Biochem Pharmacol 55:627–633
Feoktistov I, Biaggioni I (1995) Adenosine A2b receptors evoke interleukine-S secretion in human mast cells: an enprofylline-sensitive mechanism with implication for asthma. J Clin Invest 96:1979–1986
Haas HL, Selbach O (2000) Functions of neuronal adenosine receptors. Naunyn-Schmiedeberg’s Arch Pharmacodyn 362:375–381
von Lubitz DK, Ye W, McClellan J, Lin RC (1999) Stimulation of adenosine A3 receptors in cerebral ischemia. Neuronal death, recovery, or both? Ann NY Acad Sci 890:93–106
Shryock JC, Belardinelli L (1997) Adenosine and adenosine receptors in the cardiovascular system: biochemistry, physiology, and pharmacology. Am J Cardiol 79:2–10
Ramkumar V, Stiles GL, Beaven MA, Ali H (1993) The A3 adenosine receptors is the unique adenosine receptor which facilitates release of allergic mediators in mast cells. J Biol Chem 268:16887–16890
Jacobson KA (1998) Adenosine A3 receptors: novel ligands and paradoxical effects. Trends Pharmacol Sci 19:184–191
Olah ME, Ren H, Stiles GL (1995) Adenosine receptors: protein and gene structure. Arch Int Pharmacodyn Ther 329:135–150
Linden J (1994) Cloned adenosine A3 receptors: pharmacological properties, species differences and receptor functions. Trends Pharmacol Sci 15:298–306
Merighi S, Mirandola P, Varani K, Gessi S, Leung E, Baraldi PG, Tabrizi MA, Borea PA (2003) A glance at adenosine receptors: novel target for antitumor therapy. Pharmacol Ther 100:31–48
Muller CE (2003) Medicinal chemistry of A3 adenosine receptor ligands. Current Topics Med Chem 3:445–462
Moro S, Gao ZG, Jacobson, KA, Spalluto G (2006) Progress in the pursuit of therapeutic adenosine receptor antagonists. Med Res Rev 26:131–159
Baraldi PG, Cacciari B, Romagnoli R, Spalluto G, Klotz KN, Leung E, Varani K, Gessi S, Merighi S, Borea PA (1999) Pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine derivatives as highly potent and selective human A3 adenosine receptor antagonists. J Med Chem 42:4473–4478
Baraldi PG, Cacciari B, Romagnoli R, Spalluto G, Moro S, Klotz KN, Leung E, Varani K, Gessi S, Merighi S, Borea PA (2000) Pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine derivatives as highly potent and selective human A3 adenosine receptor antagonists: influence of the chain at N8 pyrazole nitrogen. J Med Chem 43:4768–4780
Baraldi PG, Cacciari B, Moro S, Spalluto G, Pastorin G, Da Ros T, Klotz KN, Varani K, Gessi S, Borea PA (2002) Synthesis, biological activity, and molecular modeling investigation of new pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine derivatives as human A3 adenosine receptor antagonists. J Med Chem 45:770–780
Baraldi PG, Cacciari B, Romagnoli R, Moro S, Ji XD, Jacobson KA, Gessi S, Borea PA, Spalluto G (2001) Fluorosulfonyl- and bis-((3-chloroethyl)amino-phenyl functionalized pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine derivatives as irreversible antagonists at the human A3 adenosine receptor: molecular modeling studies. J Med Chem 44:2735–2742
Maconi A, Pastorin G, Da Ros T, Spalluto G, Gao ZG, Jacobson KA, Baraldi PG, Cacciari B, Varani K, Borea PA (2002) Synthesis, biological properties and molecular modeling investigation of the first potent, selective and water soluble human A3 adenosine receptor antagonist. J Med Chem 45:3579–3582
Pastorin G, Da Ros T, Bolcato C, Montopoli C, Moro S, Cacciari B, Baraldi PG, Varani K, Borea PA, Spalluto G (2006) Synthesis and biological studies of a new series of 5-heteroarylcarbamoylamino-pyrazolo[4,3-e]1,2,4triazolo[1,5-c]pyrimidines as human A3 adenosine receptor antagonists. Influence of the heteroaryl substituent on binding affinity and molecular modeling investigation. J Med Chem 49:1720–1729
Moro S, Braiuca P, Deflorian F, Pastorin G, Ferrari C, Cacciari B, Baraldi PG, Varani K, Borea PA, Spalluto G (2005) Combined target-based and ligand-based drug design approach as tool to define a novel pharmacophore model of human A3 adenosine receptor antagonists: pyrazolo[4,3-e]1,2,4-triazolo[1,5-C]pyrimidine derivatives as a key study. J Med Chem 48:152–162
Pastorin G, Da Ros T, Spalluto G, Deflorian F, Moro S, Cacciari B, Baraldi PG, Gessi S, Varani K, Borea PA (2003) Pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidine derivatives as adenosine receptor antagonists. Influence of the N5 substituent on the affinity at the human A3 and A2B adenosine receptor subtypes: a molecular modeling investigation. J Med Chem 46:4287–4296
Klotz KN, Hessling J, Hegler J, Owman C, Kull B, Fredholm BB, Lohse MJ (1997) Comparative pharmacology of human adenosine receptor subtypes—characterization of stably transfected receptors in CHO cells. Naunyn-Schmiedeberg’s Arch Pharmacol 357:1–9
Moro S, Deflorian F, Spalluto G, Pastorin G, Cacciari B, et al. (2003) Demystifying the three dimensional structure of G protein-coupled receptors (GPCRs) with the aid of molecular modeling. Chem Commun 24:2949–2956
Moro S, Spalluto G, Jacobson KA (2005) Techniques: recent developments in computer-aided engineering of GPCR ligands using the human A3 adenosine receptor as an example. Trends Pharmacol Sci 26:44–51
Moro S, Deflorian F, Bacilieri M, Spalluto G (2006) Novel strategies for the design of new potent and selective human A3 receptor antagonists: an update. Curr Med Chem 13:639–645
Moro S, Bacilieri M, Deflorian F, Spalluto G (2006) G protein-coupled receptors as challenging druggable targets: insights from in silico studies. New J Chem 30:301–308
Moro S, Deflorian F, Bacilieri M, Spalluto G (2006) Ligand-based homology modeling as attractive tool to inspect GPCR structural plasticity. Curr Pharm Des 12:2175–2185
De Lean A, Hancock AA, Lefkowitz RJ (1982) Validation and statistical analysis of a computer modeling method for quantitative analysis of radioligand binding data for mixtures of pharmacological receptor subtypes. Mol Pharmacol 21:5–16
Cheng YC, Prusoff WH (1973) Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (IC50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108
OpenMosix (2004) http://www.openMosix.org
Chemical Computing Group (2006) MOE (The Molecular Operating Environment), version 2006.08 http://www.chemcomp.com
Stewart JJP (1993) MOPAC 7. Fujitsu, Tokyo
Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H et al (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289:739–745
Cornell WDCP, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force field for the simulation of proteins, nucleic acids and organic molecules. J Am Chem Soc 117:5179–5196
Levitt M (1992) Accurate modeling of protein conformation by automatic segment matching. J Mol Biol 226:507–533
Baxter CA, Murray CW, Clark DE, Westhead DR, Eldridge MD (1998) Flexible docking Using Tabù Search and an empirical estimate of binding affinity. Proteins Struct Funct Genet 33:367–382
Halgren T (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519
Acknowledgements
The molecular modeling work coordinated by S.M. has been carried out with financial support from the University of Padova, Italy. S.M. is also very grateful to Chemical Computing Group for the scientific and technical partnership.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bolcato, C., Cusan, C., Pastorin, G. et al. Pyrazolo-triazolo-pyrimidines as adenosine receptor antagonists: Effect of the N-5 bond type on the affinity and selectivity at the four adenosine receptor subtypes. Purinergic Signalling 4, 39–46 (2008). https://doi.org/10.1007/s11302-007-9058-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11302-007-9058-y