Abstract
Since the identification of CD39 and other members of the e-NTPDase (ecto-nucleoside triphosphate diphosphohydrolase) family as the primary enzymes responsible for cell surface nucleotide hydrolysis, one of their most intriguing features has been their unusual topology. The active site lies in the large extracellular region, but instead of being anchored in the membrane by a single transmembrane domain or lipid link like other ectoenzymes, CD39 has two transmembrane domains, one at each end. In this review we discuss evidence that the structure and dynamics of the transmembrane helices are intricately connected to enzymatic function. Removal of either or both transmembrane domains or disruption of their native state by detergent solubilization reduces activity by 90%, indicating that native function requires both transmembrane domains to be present and in the membrane. Enzymatic and mutational analysis of the native and truncated forms has shown that the active site can exist in distinct functional states characterized by different total activities, substrate specificities, hydrolysis mechanisms, and intermediate ADP release during ATP hydrolysis, depending on the state of the transmembrane domains. Disulfide crosslinking of cysteines introduced within the transmembrane helices revealed that they interact within and between molecules, in particular near the extracellular domain, and that activity depends on their organization. Both helices exhibit a high degree of rotational mobility, and the ability to undergo dynamic motions is required for activity and regulated by substrate binding. Recent reports suggest that membrane composition can regulate NTPDase activity. We propose that mechanical bilayer properties, potentially elasticity, might regulate CD39 by altering the balance between stability and mobility of its transmembrane domains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
While the existence of divalent cation dependent nucleotidase activities on the outer surface of the plasma membrane of animal cells has been known for over 50 years [1, 2], the molecular identification of the relevant proteins began in 1996 when potato apyrase, an enzyme with properties similar to those of the plasma membrane nucleotidases, was cloned [3]. Several members of this class of proteins have since been cloned [4–8]. All family members share a set of short sequences termed apyrase conserved regions (ACRs), and the mammalian family members are collectively referred to as ecto-nucleoside triphosphate diphosphohydrolases (e-NTPDases). Of the eight members of this family, all have two transmembrane domains except for NTPDases 5 and 6. NTPDase1 is CD39. NTPDases 1, 2, 3 and 8 are located in the plasma membrane, while the UDPase and a similar protein cloned from pancreatic tumor cells, LALP70 [9] both called NTPDase 4, are in the Golgi membranes and lysosomal vacuoles. NTPDase 7 is present in intracellular membranes [7].
Ecto-enzymes, for instance 5′-nucleotidase and alkaline phosphatase, are generally anchored to the membrane by a single transmembrane domain or lipid link [10]. Why do the plasma membrane bound NTPDases in animal cells have two transmembrane domains, one at each end of the molecule? Most proteins with this structure are channels (K channels, mechanosensitive channels, purinergic receptors (P2X) and epithelial Na channels [11]). In this review we discuss the development of the idea that the transmembrane domains of the NTPDases, in additional to simply anchoring the protein in the membrane, play an integral role in the function and regulation of the enzymes.
Native CD39 activity requires two transmembrane domains
The original clue to a connection between the transmembrane domains and active site function of the cell surface NTPDases was the loss of activity upon detergent solubilization. Pre-cloning attempts to study the proteins by purifying them from native membranes had been frustrated by the fact that nearly every detergent used for extraction abolished most of the enzymatic activity [12]. Following the cloning of the soluble potato apyrase [3] and the subsequent identification of CD39 as the major membrane bound enzyme responsible for cell surface apyrase activity [13], two things became apparent. First, CD39 had the unusual feature of being linked to the membrane by two transmembrane domains flanking its extracellular active site [14], in contrast to the lipid link or single transmembrane domain observed for most other ecto-enzymes [10]. Second, despite conservation of the ACRs [3] thought to constitute the nucleotide binding site, detergent sensitivity was a property of the membrane bound CD39 but not of the soluble potato apyrase [15] or of the related soluble T. gondii NTPases [16]. These two pieces of information led Wang and Guidotti to ask whether disruption of transmembrane domain interactions is the mechanism for detergent effects, and thus whether the two transmembrane domains are required for enzymatic activity.
The answer to both questions was yes. Wang et al. [15] systematically deleted the amino terminal (TM1), carboxy terminal (TM2) or both transmembrane domains of CD39 and compared the activity and detergent sensitivity of each truncated protein with native CD39. Truncation of both transmembrane domains resulted in a catalytically active secreted soluble protein that, like the naturally occurring soluble family members, was resistant to detergent. Furthermore, its activity was about 90% lower than native CD39 and approximately equal to that of detergent solubilized CD39. Thus, removal of the transmembrane domains both mimicked and precluded the detergent effect, supporting the hypothesis that the transmembrane domains are required for activity and that detergents disrupt activity via them rather than by binding to the active site. While this result left open the possibility that removal from the membrane might be the crucial factor, the single truncation versions of CD39 narrowed the focus to structural features of the transmembrane domains themselves. Deletion of either TM1 or TM2 resulted in a catalytically active enzyme that was targeted to and rooted in the plasma membrane; nevertheless, both proteins were resistant to detergents and had low enzymatic activity similar to that of the double truncated enzyme. These studies made clear that native CD39 activity requires both TM1 and TM2 to be present and in the membrane.
Wang et al. [15] further proposed that not only the presence and membrane insertion of the transmembrane domains but also the interactions between them are required for native activity. Glutaraldehyde crosslinking as well as sucrose density gradient sedimentation in the presence of detergents showed that the protein formed oligomers, that detergents that reduced activity dissociated the oligomers, and that the soluble extracellular domain lacking both transmembrane domains existed only as a monomer. Furthermore, the one detergent that had only a small effect on activity, digitonin, preserved oligomers. Together these data suggested that the transmembrane domains mediate oligomer formation and might contribute to enzymatic function by this means. However, the correlation between oligomerization and native activity was not exact; both the TM1 and TM2 single truncated proteins also formed oligomers despite their detergentlevel activity, suggesting that a specific set of interactions involving both TM1 and TM2 might underlie their influence on the active site.
While the nature and importance of oligomerization or of any particular interactions between the transmembrane domains remained uncertain, the work of Wang et al. established that understanding the relationship between the transmembrane domains and the active site was critical to understanding CD39 function. In the following sections we discuss first the development of the idea that CD39 can exist in at least two different functional states characterized by distinct quantitative and qualitative enzymatic properties, each with distinct physiological consequences, depending on the state of its transmembrane domains. We then discuss experiments and questions concerning what exactly the transmembrane domains do at the structural and dynamic level and how they might enable CD39 function to be regulated by its membrane environment.
Transmembrane domains govern switching between mechanistically and enzymatically distinct forms of the active site
The Wang et al. experiments revealed that disrupting the transmembrane domains significantly reduced the rate of nucleotide hydrolysis, but due to the high specific activity of native CD39 the retention of 10% activity still left the disrupted forms with a respectable enough hydrolysis rate to be studied in their own right. Through the experiments described below it has become clear that the disrupted forms are not simply a weakened version of native CD39. Not only total activity but also the hydrolysis mechanisms, role of ACR1, discrimination between substrates, and presence or absence of intermediate ADP release during ATP hydrolysis are all at the mercy of the transmembrane domains. For all cases looked at, the various properties fall into line one way for native CD39 and the alternative way for the soluble double truncated, single TM1 and TM2 truncated, and detergent solubilized forms, suggesting that the active site can exist in two distinct states depending on the presence or absence of native transmembrane domain organization.
The insight into these differences originated from initially transmembrane domain-blind efforts to understand the hydrolysis mechanism and role of the ACRs of CD39 as well as the basis for the different substrate specificities of various NTPDase family members. While a specific active site had not been determined for any family members, attention was focused on the five ACRs [3, 17], particularly on ACR1 and ACR4 due to their homology with the crystallographically identified phosphate binding loops of the actin/hsp70/hexokinase superfamily [3, 18]. Mutagenesis of completely conserved residues in these regions was found to abolish both ATPase and ADPase activity, consistent with their constituting the active site for both ATP and ADP hydrolysis [19–22]. However, ACR sequence comparisons among family members with different substrate specificities, particularly between apyrases and ATPases, offered no clues to how the enzymes discriminate between ATP and ADP; in many cases the ACRs of apyrases and ATPases were identical [16, 23, 24].
On the other hand, the two-transmembrane domain apyrases and ATPases known at the time did exhibit conserved sequence differences in their transmembrane domains [23, 24]. Added to this was the somewhat obscure fact that two residues in the putative β-phosphate binding loop ACR1 correlated with the presence or absence of transmembrane domains [23]. Among the cloned family members, most of those with two transmembrane domains had a histidine at the position corresponding to H59 in CD39 and asparagine, serine, or alanine in the position corresponding to N61, while all with zero or one transmembrane domain had glycine/serine and arginine at these positions. Given the backdrop of the recent discovery that the transmembrane domains influence active site function, these two sets of correlations suggested a potential relationship among the role of ACR1, substrate specificity, and the contribution of the transmembrane domains to ATPase and ADPase activities.
Mutation of these ACR1 residues in both native CD39 and its truncated and detergent solubilized versions revealed not only a point of distinction between ATP and ADP hydrolysis mechanisms but also that the enzyme uses different hydrolysis mechanisms depending on the state of its transmembrane domains [23]. Substituting G or S for H59 in native CD39 abolished approximately 95% of the ATPase activity but less than 50% of the ADPase activity and thus converted CD39 from an apyrase with a 1.4:1 ATPase:ADPase rate ratio to primarily an ADPase with a 6:1 (for H59G) or 8:1 (H59S) ADPase:ATPase ratio. However, the effect of these substitutions was completely different in the truncated and solubilized backgrounds. When introduced into the soluble form of CD39 lacking both transmembrane domains, H59G and H59S substitution had no effect whatsoever on ATPase activity and caused a twofold increase in ADPase activity. The result was identical for the TM1 and TM2 single truncated forms and for the detergent solubilized full length protein. Substitution of N61 with R had a more modest effect on the ATPase:ADPase ratio, changing it only to a 2:1 ADPase:ATPase ratio, but similarly reduced activity in the native context and had no effect in the truncated and solubilized backgrounds. These results were in contrast to mutation of completely conserved residues in ACR1 as well as in ACRs 2, 3, and 4, which abolished both ATPase and ADPase activity in both the native and truncated contexts [19–22].
Thus ATP hydrolysis was shown to occur through a mechanism characterized by a critical dependence on H59 in the native enzyme while occurring through a different mechanism that completely bypasses this residue when the transmembrane domains are disrupted. The fact that other residues in ACR1 were equally critical in both contexts suggested that the active site does not relocate; instead the way the substrate is hydrolyzed by the same active site appeared to change. The same story turned out to be true for the distinction between ATP and ADP hydrolysis mechanisms in the native context; while both substrates use the same active site, ATP was notably more vulnerable to H59 substitution than was ADP. The fact that the same residue represented a pivot point distinguishing between native and non-native ATP hydrolysis mechanisms on one hand, and between ATP and ADP hydrolysis mechanisms on the other hand, added up to the conclusion that the transmembrane domains, at least in part via H59, are responsible for the distinction between the native ATP and ADP hydrolysis mechanisms.
This conclusion suggested that the transmembrane domains might also regulate substrate specificity. Support for this hypothesis was provided by a set of experiments comparing CD39 with CD39L1 (NTPDase2), a family member that like CD39 has two transmembrane domains and loses about 90% of its activity when solubilized but which hydrolyzes primarily ATP [25]. Despite their different substrate specificities in the native state, upon solubilization or double truncation each enzyme arrived at an intermediate ATPase:ADPase ratio of 3.5:1. Thus, removal or disruption of transmembrane domains not only lowered the activity of both enzymes but removed the distinction between the apyrase and the ATPase and left them both with the same intermediate ATPase:ADPase ratio. This discovery supported the idea that the extracellular domains of the membrane bound apyrase and ATPase have the same intrinsic substrate specificity and that the state of their transmembrane domains determines whether they remain in this basic form or assume their native identities as an apyrase or ATPase.
Further studies [25] suggested that the key to how the transmembrane domains bring about this distinction might be their ability to confer different ADP hydrolysis mechanisms on the two enzymes. Similarly to CD39, CD39L1 was found to use different ATP hydrolysis mechanisms in its native and soluble states. Substituting H50, the equivalent of H59 in CD39, with G or S reduced the ATPase activity of native CD39L1 by approximately 90% but had no significant effect on either ATPase or ADPase activity in CD39L1 solubilized in detergent or lacking both transmembrane domains. However, unlike in CD39, in native CD39L1 these substitutions had the same effect on ADPase activity as on ATPase activity; they reduced ADPase activity by 90%, so that rather than changing the enzyme into an ADPase as in CD39, H50 substitution caused no change in native CD39L1 substrate specificity. These results suggested that while the state of the transmembrane domains causes both enzymes to switch between H50/59 dependent and independent ATP hydrolysis mechanisms, the native transmembrane domain state also brings about a difference in ADP hydrolysis mechanisms between the apyrase and the ATPase. While for both enzymes native transmembrane domain organization causes an overall increase in ADPase activity relative to the solubilized state, the native ADP hydrolysis mechanism is critically dependent on H50 in CD39L1 but relatively independent of H59 in CD39. Neither enzyme requires H50/59 for ADP hydrolysis in the solubilized state. These data suggested that the native transmembrane state confers the distinct substrate specificities on the apyrase and ATPase by conferring distinct ADP hydrolysis mechanisms.
Concurrently with this work on hydrolysis mechanisms and substrate specificity, it was also discovered that the transmembrane domains determine whether or not ADP is released from the active site as an intermediate during ATP hydrolysis. In all cases CD39 was known to hydrolyze one phosphate at a time; pyrophosphate was not released under any circumstances. Nevertheless, Heine et al. [26] showed that when CD39 hydrolyzed ATP to completion, an ADP intermediate was never observed in the process; only a decrease in ATP and a subsequent increase in AMP occurred. In contrast, Chen and Guidotti [27] found that soluble CD39 lacking both TM1 and TM2 did exhibit intermediate ADP release; ATP disappearance was followed by the appearance of ADP, which subsequently decreased with a concomitant buildup of AMP. These data suggested that in native CD39, the ATP and ADP hydrolysis mechanisms are such that ATP and ADP hydrolysis can occur in succession without dissociation of ADP, while in soluble CD39 the two mechanisms are such that ADP as product must dissociate and rebind to the active site as substrate. Such a scenario would be consistent with the above mentioned finding that the ATP and ADP hydrolysis mechanisms are distinguished by their dependence and independence of H59 in native CD39 but have no such distinction in soluble CD39; the ability to use the same active site differently may be key to the ability to hydrolyze ADP without releasing and rebinding it. Consistent with this idea, Heine et al. [26] observed an intermediate buildup of ADP prior to production of AMP with native CD39L1, which, as described above, has ATP and ADP hydrolysis mechanisms that use H50 to the same extent and may therefore lack the distinction between ATP and ADP hydrolysis mechanisms required for successive hydrolysis of both phosphates.
Mobility of transmembrane helices regulates and is regulated by the active site
Together, the studies described above revealed that the answer to several of the physiologically important questions concerning CD39 — its total activity, mechanism, substrate specificity, whether ADP is produced upon ATP hydrolysis — is that it depends on the state of the transmembrane domains. To the extent studied, the case looks similar for CD39L1 and could potentially apply to other two transmembrane domain NTPDase family members. The question now turns to the transmembrane domains themselves: What specifically about them is responsible for their effects on the active site, and, ultimately, how might they change within the context of the natural membrane environment and potentially manifest the different forms of the enzyme?
In theory, any of a broad range of structural features might underlie CD39’s requirement for both TM1 and TM2 to be present and in the membrane. Independent of any interactions between them, they might serve to position the adjacent ends of the extracellular domain at a fixed distance from the membrane, thus stabilizing the structure by eliminating one degree of freedom in the direction perpendicular to the membrane. They might also interact to limit both the lateral and rotational degrees of freedom of the ends of the extracellular domain relative to each other. Either case would be likely to have a direct influence on the active site, since ACR1 and ACR5 are only about 10 to 20 amino acids from TM1 and TM2, respectively. With or without intramolecular interactions between TM1 and TM2, the transmembrane domains might work by stabilizing oligomers, as proposed by Wang et al. [15]. Oligomer stabilization might be achieved nonspecifically by reducing the effective concentration of CD39 molecules by limiting their movement to the two dimensions of the membrane rather than the three in solution; if either of the first two cases is true then specific intermolecular transmembrane domain interactions would not necessarily have to be invoked to explain the requirement for both TM1 and TM2. Alternatively, TM1, TM2, or both might work by interacting intermolecularly in various combinations to produce an oligomer with a specific orientation of subunits. Such interactions might occur in the presence or absence of intramolecular TM1–TM2 interactions.
Most discussion of the role of the transmembrane domains has revolved around oligomerization due to the fact that CD39 as well as other family members forms oligomers in various crosslinking, sedimentation, and native gel experiments [15, 28, 29]. Nevertheless, the jury has remained out on whether oligomerization is essential to native activity because no experiments have been able to demonstrate a direct causal relationship. Oligomerization per se does not correlate with native CD39 activity; both the TM1 and TM2 single truncated constructs [15] as well as full length CD39 solubilized in cholate form oligomers but have the same enzymatic properties as soluble CD39, indicating oligomerization is not sufficient for native properties. To date no experiment has been able to distinguish whether oligomerization is necessary for native activity because all experiments that can be shown to dissociate oligomers also potentially disrupt intramolecular TM1–TM2 interactions or remove CD39 from the membrane.
A more comprehensive picture of both intra- and intermolecular transmembrane domain interactions and how they relate to activity was recently developed via oxidative crosslinking of cysteines introduced into TM1 and TM2 [30]. The technique is based on the principle that if cysteines are introduced in pairs, only pairs that are oriented toward each other and are within the length of a disulfide bond can form disulfides upon addition of an oxidizing agent. Since oxidation can occur within the membrane, CD39 transmembrane domain interactions were analyzed and correlated with activity and substrate binding in the native membrane environment.
Crosslinking of cysteines substituted one at a time along TM1, along TM2, and at various pairs of TM1 and TM2 positions revealed that the transmembrane domains do in fact interact both within and between CD39 molecules. Strong intramolecular TM1–TM2 interactions as well as intermolecular TM1–TM1′ and TM2–TM2′ interactions were observed near the extracellular side of the membrane, with weaker interactions near the cytoplasmic side. This evidence for transmembrane helix packing, particularly in the region closest to the active site, supported the idea that specific interactions might underlie the transmembrane domains’ role in enzyme function. However, the two helices turned out to exhibit a high degree of flexibility concerning which helix faces constituted the interaction interfaces; under crosslinking conditions used in several other membrane proteins to identify interacting helix faces [31–33], complete disulfide formation occurred between all faces of TM1 and TM2 within a molecule as well as between all faces of TM1 and TM1′ and TM2 and TM2t’ of different molecules. The failure of multiple cysteines per helix to increase the size of the crosslinked oligomers ruled out the possibility of a large array of subunits or of subunits randomly colliding with no specificity. This result, together with the multiple intramolecular TM1–TM2 interfaces, indicated that while the transmembrane domains do interact, each helix exhibits a high degree of rotational mobility, presumably within the packed structure. In nearly all cases TM1–TM2 disulfide formation precluded TM1YTM1′ or TM2′–TM2′ crosslinking, indicating that even with rotational mobility taken into account, interactions within a subunit are stronger than those between subunits.
Despite the high degree of mobility, distinct helix faces could be discerned by differences in temperature dependence of disulfide formation at different positions. At low temperatures crosslinking was limited primarily to positions along one side of each helix, suggesting that the helices do have specific favored interaction surfaces rather than complete randomness. However, locking the helices in this orientation by forming disulfides at the positions most readily crosslinked at low temperature neither increased activity nor protected the enzyme from loss of activity in response to detergent. Rather, crosslinking at any position, both within and between subunits, resulted in loss of activity comparable to the loss induced by solubilizing or by removing the transmembrane domains altogether. Such was the case for an exhaustive combination of TM1–TM2 pairs representing all combinations of helical faces as well as a variety of relative depths in the membrane, as well as for every possible TM1–TM1′ and TM2–TM2′ linkage between subunits.
The loss of activity caused by locking the helices in any single orientation revealed, first, that both intra- and intermolecular interactions between the transmembrane domains are a crucial factor in their regulation of active site function; despite the high degree of mobility, the enzyme cannot function with its transmembrane domains in random orientations. At the same time, these results disfavored the idea that the transmembrane domains work by stabilizing one optimal orientation; rather the data suggested that they must be able to move relative to each other during the process of nucleotide binding and hydrolysis. Such movements may be necessary within and between subunits.
Further supporting a relationship between active site and transmembrane domain dynamics was the observation that ATP binding regulates transmembrane helix crosslinking. In the absence of ATP, as described above, all positions could be crosslinked at physiological temperatures; however in the presence of ATP crosslinking at physiological temperature was limited to the same helical pattern seen without ATP at low temperatures. The effect was mimicked by another substrate, ADP, but not by AMP or phosphate. These results indicated that substrate binding stabilizes the transmembrane helices in a specific arrangement and thus directly revealed a structural interdependence between active site and transmembrane domain structure and dynamics. While the specifics of how exactly different stages of nucleotide binding and hydrolysis might correlate with different transmembrane domain arrangements remain to be worked out, these results suggested that changes in active site structure during the process of hydrolysis may be tied to changes in transmembrane domain arrangement.
Thus, in addition to specific interactions between the transmembrane domains, the ability of the transmembrane helices to undergo dynamic motions may underlie their regulation of enzymatic function. In these studies, mobility itself represents a parameter that can both regulate and be regulated by the active site. While locking the transmembrane helices in place limits mobility, removing or solubilizing them may represent the opposite extreme; lack of interactions between them may provide too much freedom to the active site and reduce activity by uncoupling active site and transmembrane helix movements (Figure 1). The balance between stability and flexibility may therefore be a critical feature to examine when asking how the transmembrane domains might change in the context of the native membrane environment to regulate potential shifting between different states of the enzyme.
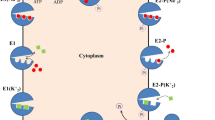
Cartoon of the structure of CD39. The region labeled PM represents the lipid bilayer of the plasma membrane. TM1 and TM2 are the N-terminal and the C-terminal transmembrane domains, respectively. The hourglass structures attached to TM1 and TM2 represent ACR1 and ACR5, respectively; they are part of the extracellular domain of CD39 (cylinder). The dynamic interactions between TM1 and TM2 in wild type (WT) CD39 may position ACR1 and ACR5 so that the configurations and mobility of the enzymatic active site are optimal and the specific activity is maximal. Removal of TM1 (ΔTM1), TM2 (ΔTM) or both transmembrane domains (ΔTM1 + ΔTM2) eliminates the interactions between TM1 and TM2 and would be expected to disrupt the alignment of ACR1 and ACR5 and of the enzymatic active site to bring about a loss of specific activity.
Native CD39 may sense bilayer mechanical properties via its transmembrane helices
While understanding how the transmembrane domains contribute to activity provides insight into how CD39 works, it remains unknown to what extent the transmembrane helices themselves may be regulated by their environment. Membrane proteins in general vary in the extent to which they can be regulated by membrane context, and the factors that determine how much transmembrane helix behavior is determined by intrinsic sequence properties and how much by the surrounding lipids are not well understood. To date the question of whether CD39 can be converted within native cell membranes to exhibit the mechanistic properties of the solubilized and truncated forms has not been examined in detail. However, a recent report provides compelling evidence that such regulation by membrane environment may indeed occur. Papanikolaou et al. [34] show that removing cholesterol from membranes prepared from CD39-expressing cells reduces ATPase activity to the same extent as does solubilization. Activity loss is correlated with the amount of cholesterol remaining in the membrane. While the low activity may or may not indicate the presence of different hydrolysis mechanisms and enzymatic properties observed for the solubilized and truncated forms of CD39, their finding that readdition of cholesterol to the membrane restores activity indicates that the protein is not simply denatured.
Regulation of activity by cholesterol content presumably occurs via changes in the transmembrane domains, but understanding how they sense cholesterol content and relay it to the active site requires comparison of their structure and dynamics in the presence of different amounts of cholesterol. In addition, examining a broad assortment of reagents and other means for altering membrane structure is necessary to gain insight into the mechanical features of the membrane that underlie such changes. The demonstration by Papanikolaou et al. [34] that adding cholestenone to cholesterol depleted membranes does not restore CD39 activity suggests that some aspect of cholesterol other than its effect on membrane fluidity is required for native transmembrane helix properties but leaves open the question of whether cholesterol acts through direct binding to CD39 or via general changes in membrane properties other than fluidity.
Recent studies by Mukasa et al. [35] on two related two transmembrane domain NTPDases, NTPDase2 and 8, suggest that at least these family members may be regulated by general properties of the membrane. NTPDase2 activity is decreased by concentrations of NP40 that are not expected to solubilize it, as well as by high temperature. NTPDase8 activity is resistant to NP40 and increased by high temperature, but inhibited by the combination of NP40 and high temperature. Exchanging transmembrane domains between the two enzymes demonstrates that these effects are mediated at least in part via the membrane. While comparisons among family members are complicated by the fact that they differ in their transmembrane domain sequences and detergent sensitivity, these experiments suggest that NTPDases may be regulated by a broad array of structurally unrelated membrane perturbing factors.
Mukasa et al. [35] propose that their results may be explained by changes in oligomer stability due to changes in membrane fluidity; however, as discussed above, fluidity does not explain the effect of cholesterol on CD39. The different enzymes may sense different features of the membrane environment. Alternatively, the various perturbing methods may change a common mechanical feature other than fluidity. Along these lines, Lundbaek et al. [36] have identified bilayer elasticity as a membrane property that is altered by a variety of structurally unrelated compounds as well as by cholesterol depletion and which can explain the common effects of all these treatments on the function of gramicidin and cardiac sodium channel function. Changes in elasticity change the energy required for local membrane deformation associated with a protein conformational change and can therefore modify the total energy barrier between different transmembrane domain conformations. If, as proposed above, the balance between stability and mobility is a key feature of the interplay between the transmembrane domains and active site of CD39, anything that changes bilayer elasticity might change activity by altering this balance. In particular, changes that increase elasticity, such as detergents and cholesterol depletion, might increase mobility by lowering the barriers among different conformations and thus uncouple transmembrane domain and active site dynamics as proposed for truncated and solubilized CD39.
Physiologically, extracellular nucleotide concentrations are directly linked to a number of mechanical and biochemical processes that change the properties of the membrane (reviewed in [37–39]). ATP release is a primary response to mechanical pressure and shear stress as well as to changes in osmotic pressure; in the latter case the resulting extracellular ATP is required to restore osmotic balance. Inflammatory processes and oxidative stress, both of which rely on signaling by extracellular nucleotides, trigger release of unsaturated fatty acids and lysophosphatidylcholine, both of which can insert in the membrane and change its curvature and elasticity. High cholesterol levels are associated with blood clotting, which is modulated by extracellular nucleotides. In each of these cases the ability to modulate the hydrolysis rate, substrate specificity, and intermediate ADP release of CD39 according to the mechanical state of the membrane would provide a direct feedback mechanism between cellular conditions that affect the membrane and the extracellular nucleotide signaling processes that respond to them. Thus, the presence of two transmembrane domains might be a key to instantaneously tailoring extracellular nucleotide signaling to the state of the cell.
Preliminary work suggests that CD39 may have functions beyond apyrase activity, also potentially mediated by its transmembrane domains. Bodas et al. [40] have shown that CD39 can act as an ATP channel, while Wu et al. [41] have found that the short cytoplasmic region of the protein binds ran binding protein and thus potentially plays a role in intracellular signaling. If CD39 is confirmed to serve as a transporter of nucleotides or of signals across the membrane, the structural and dynamic regulation by and of the two transmembrane domains may underlie an interdependence among the active site, the bilayer, and the internal state of the cell.
Abbreviations
- e-NTPDase:
-
ecto-nucleoside triphosphate diphosphohydrolase
- ACR:
-
apyrase conserved region
- TM1:
-
transmembrane domain 1
- TM2:
-
transmembrane domain 2
References
Rothstein A, Meier RC, Scharff TG. Relationship of cell surface to metabolism: IX. Digestion of phosphorylated compounds by enzymes located on the surface of intestinal cells. Am J Phys 1953; 173: 41–6.
Ziganshin AU, Hoyle CHV, Burnstock G. Ecto-enzymes and metabolism of extracellular ATP. Drug Dev Res 1994; 32: 134–46.
Handa M and Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum). Biochem Biophys Res Commun 1996; 218: 916–23.
Wang TF, Guidotti G. Golgi localization and functional expression of human uridine diphosphatase. J Biol Chem 1998; 273: 11392–11399.
Zimmermann H, Beaudoin AR, Bollen M et al. Proposed nomenclature for two novel nucleotide hydrolysing enzyme families expressed on the cell surface. In vanDuffel L, Lemmens R (eds): Ecto-ATPase and Related Nucleotidases, Maastricht: Shaker, 2000, 1–8.
Braun N, Fengler, S, Ebeling C et al. Sequencing, functional expression and characterization of rat NTPDase6, a nucleoside diphosphatase and novel member of the ecto-nucleoside triphosphate diphosphohydrolase family. Biochem J 2000; 351: 467–639.
Shi JD, Kukar T, Wang CY et al. Molecular cloning and characterization of a novel mammalian endo-apyrase (LALP1). J Biol Chem 2001; 276: 17474–8.
Bigonnesse F, Levesque SA, Kukulski F et al. Cloning and characterization of mouse nucleoside triphosphate diphosphohydrolase-8. Biochemistry 2004; 43: 5511–9.
Biederbick A, Rose S, Elsasser H-P. A human intracellular apyrase-like protein, LALP70, localizes to lysosomal/autophagic vacuoles. J Cell Sci 1999; 112: 2473–84.
Semenza G. Anchoring and biosynthesis of stalked brush border membrane proteins: Glycosidases and peptidases of enterocytes and renal tubuli. Annu Rev Cell Biol 1986; 2: 255–313.
North RA. Families of ion channels with two hydrophobic segments. Curr Opin Cell Biol 1996; 8: 474–83.
Plesner L. Ecto-ATPases: Identities and functions. Int Rev Cytol 1995; 158: 141–214.
Wang TF, Guidotti G. CD39 is an ecto-(Ca2+, Mg2+)-apyrase. J Biol Chem 1996; 271: 9898–901.
Maliszewski CR, Delespesse GJ, Schoenborn MA et al. The CD39 lymphoid cell activation antigen. Molecular cloning and structural characterization. J Immunol 1994; 153: 3574–83.
Wang TF, Ou Y, Guidotti G. The transmembrane domains of ectoapyrase (CD39) affect its enzymatic activity and quaternary structure. J Biol Chem 1998; 273: 24814–21.
Bermudes D, Peck KR, Afifi MA et al. Tandemly repeated genes encode nucleoside triphosphate hydrolase isoforms secreted into the parasitophorous vacuole of Toxoplasma gondii. J Biol Chem 1994; 269: 29252–60.
Schulte am Esch J, Sevigny J, Kaczmarek E et al. Structural elements and limited proteolysis of CD39 influence ATP diphosphohydrolase activity. Biochemistry 1999; 38: 2248–58.
Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA 1992; 89: 7290–94.
Handa M. PhD Thesis. Cambridge: Harvard University 1997.
Smith TM, Kirley TL. Site-directed mutagenesis of a human brain ecto-apyrase: Evidence that the E-type ATPases are related to the actin/heat shock 70/sugar kinase superfamily. Biochemistry 1999; 38: 321–29.
Drosopoulos JH, Broekman MJ, Islam N et al. Site-directed mutagenesis of human endothelial cell ecto-ADPase/soluble CD39: Requirement of glutamate 174 and serine 218 for enzymatic activity and inhibition of platement recruitment. Biochemistry 2000; 39: 6936–43.
Yang F, Hicks-Berger CA, Smith TM, Kirley TL. Site-directed mutagenesis of human nucleoside triphosphate diphosphohydrolase 3: The importance of residues in the apyrase conserved regions. Biochemistry 2001; 40: 3943–50.
Grinthal A, Guidotti G. Substitution of His59 converts CD39 apyrase into an ADPase in a quaternary structure dependent manner. Biochemistry 2000; 39: 9–16.
Chadwick BP, Frischauf AM. The CD39-like gene family: Identification of three new human members (CD39L2, CD39L3, and CD39L4), their murine homologues, and a member of the gene family from Drosophila melanogaster. Genomics 1998; 50: 357–67.
Grinthal A, Guidotti G. Transmembrane domains confer different substrate specificites and adenosine diphosphate hydrolysis mechanisms on CD39, CD39L1, and chimeras. Biochemistry 2002; 41: 1947–56.
Heine P, Braun N, Heilbronn A, Zimmermann H. Functional characterization of rat ecto-ATPase and ecto-ATP diphosphohydrolase after heterologous expression in CHO cells. Eur J Biochem 1999; 262: 102–7.
Chen W, Guidotti G. Soluble apyrases release ADP during ATP hydrolysis. Biochem Biophys Res Comm 2001; 282: 90–5.
Failer BU, Aschrafi A, Schmalzing G, Zimmermann, H. Determination of native oligomeric state and substrate specificity of rat NTPDase1 and NTPDase2 after heterologous expression in Xenopus oocytes. Eur J Biochem 2003; 270: 1802–9.
Knowles AF, Chiang WC. Enzymatic and transcriptional regulation of human ecto-ATPase/E-NTPDase 2. Arch Biochem Biophys 2003; 418: 217–27.
Grinthal A, Guidotti G. Dynamic motions of CD39 transmembrane domains regulate and are regulated by the enzymatic active site. Biochemistry 2004; 43: 13849–58.
Hughson AG, Lee GF, Hazelbauer GL. Analysis of protein structure in intact cells: Crosslinking in vivo between introduced cysteines in the transmembrane domain of a bacterial chemoreceptor. Protein Sci 1997; 6: 315–22.
Hamdan FF, Ward SDC, Siddiqui NA et al. Use of an in situ disulfide crosslinking strategy to map proximities between amino acid residues on transmembrane domains I and VII of the M3 muscarinic acetylcholune receptor. Biochemistry 2002; 41: 7647–58.
Bunn MW, Ordal GW. Transmembrane organization of the Bacillus subtilis chemoreceptor McpB deduced by disulfide crosslinking. J Mol Biol 2003; 331: 941–9.
Papanikolaou A, Papfotika A, Murphy C et al. Cholesterol-dependent lipid assemblies regulate the activity of the ecto-nucleotidase CD39. J Biol Chem 2005; 280: 26406–14.
Mukasa T, Lee Y, Knowles AF. Either the carboxyl- or the amino-terminal region of the human ecto-ATPase (E-NTPDase 2) confers detergent and temperature sensitivity to the chicken ecto-ATP-diphosphohydrolase (E-NTPDase 8). Biochemistry 2005; 44: 11160–70.
Lundbaek JA, Birn P, Hansen AJ et al. Regulation of sodium channel function by bilayer elasticity: The importance of hydrophobic coupling. Effects of micelle-forming amphiphiles and cholesterol. J Gen Physiol 2004; 123: 599–621.
Schweibert EM, Zsembery A. Extracellular ATP as a signaling molecule for epithelial cells. Biochim Biophys Acta 2003; 1615: 7–32.
Brake AJ, Julius D. Signaling by extracellular nucleotides. Annu Rev Cell Dev Biol 1996; 12: 519–41.
Burnstock G. In Turner HT, Weisman GA, Fedan JS (eds): The P2 Nucleotide Receptors. Totowa: Humana, 1998; 3–42.
Bodas E, Aleu J, Pujol G et al. ATP crossing the cell plasma membrane generates an ionic current in Xenopus oocytes. J Biol Chem 2000; 272: 20268–73.
Wu Y, Sun X, Keiichi Enjyoji K, Robson SC. CD39 Interacts with Ran Binding Protein M (RanBPM) to directly modulate Ras activation and cellular proliferation in liver regeneration following partial hepatectomy. Hepatology 2004; 135; 40, 222.
Acknowledgement
This work was supported by grant HL08893 from the National Institutes of Health to GG.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Grinthal, A., Guidotti, G. CD39, NTPDase 1, is attached to the plasma membrane by two transmembrane domains. Why?. Purinergic Signalling 2, 391–398 (2006). https://doi.org/10.1007/s11302-005-5907-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11302-005-5907-8