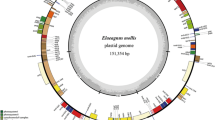
Abstract
The genus Rosa comprises more than 150 species spread across three subgenera, Hesperhodos, Hulthemia, and Rosa, most of which have high economic and ecological values. Here, we report 31 complete plastomes that belong to the genus Rosa, with the aim of better understanding the evolution and divergence of genes of the plastome in this genus. A comparative analysis was conducted to characterize the chloroplast genomes of 12 taxa that cover all the sections in the three subgenera of Rosa. Further, complete chloroplast genome sequences revealed six hotspots of nucleotide polymorphism, including five intergenic regions and one coding sequence. In addition, a pairwise analysis revealed that R. stellata and R. berberifolia have the highest average genetic distances (Da) and nucleotide divergence (Dxy) compared with other species. Moreover, the lowest Da and Dxy was observed between R. gallica and R. canina, followed by R. multiflora and R. chinensis var. spontanea. The phylogenetic relationships within Rosa inferred from the 44 chloroplast genomes revealed the R. subg. Hesperhodos is the clade that diverged the earliest. Its successive clades were identified as R. subg. Huithemia and R. sect. Pimpinellifolia. The phylogenomic analysis also revealed rapid simultaneous diversification within the Rosa subgenus. Significant increases in Pi and d N for ycf1, d N/d S for ycf2 were observed across the genus. Finally, we found that most RNA editing sites identified in the genus are section-specific, suggesting that the subgenera or sections have a self-evolving lineage. Taken together, the plastome information is valuable for species identification, phylogenetic studies, molecular genetics and breeding Rosa species.










Similar content being viewed by others
Change history
22 July 2022
Handling editor name correction.
References
Allen GC, Floresvergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethy lammonium bromide. Nat Protoc 1:2320–2325
Barrie FR (2006) Report of the general committee: 9. Taxon 55:795–800
Bendahmane M, Dubois A, Raymond O, Bris ML (2013) Genetics and genomics of flower initiation and development in roses. J Exp Bot 64:847–857
Bruneau A, Starr JR, Joly S (2007) Phylogenetic relationships in the genus Rosa: new evidence from chloroplast DNA sequences and an appraisal of current knowledge. Syst Bot 32:366–378
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chen MR, Zhang C, Gao XF (2019) The complete chloroplast genome sequence of Rosa pricei (Rosaceae). Mitochondrial DNA B 4:1918–1919
Chen X, Liu YL, Sun JH, Wang L, Zhou SL (2019) The complete chloroplast genome sequence of Rosa acicularis in Rosaceae. Mitochondrial DNA B 4:1743–1744
Cui WH, Du XY, Zhong MC, Fang W, Suo ZQ, Wang D, Dong X, Jiang XD, Hu JY (2022) Complex and reticulate origin of edible roses (Rosa, Rosaceae) in China. Hortic Res 9. https://doi.org/10.1093/hr/uhab051
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. Plos One 5:e11147
Debener T, Linde M (2009) Exploring complex ornamental genomes: the rose as a model plant. Crit Rev Plant Sci 28:267–280
Debray K, Marie-Magdelaine J, Ruttink T, Clotault J, Foucher F, Malécot V (2019) Identification and assessment of variable single-copyorthologous (SCO) nuclear loci for low-level phylogenomics: a case study in the genus Rosa (Rosaceae). BMC Evol Biol 19:152
Debray K, Le Paslier M-Ch, Bérard A, Thouroude T, Michel G, Marie-Magdelaine J, Bruneau A, Foucher F, Malécot V (2021) Unveiling the patterns of reticulated evolutionary processes with phylogenomics: hybridization and polyploidy in the genus Rosa. Syst Biol. https://doi.org/10.1093/sysbio/syab064
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45:e18
Fan JL (2020) Plant genomics. Science Press, Beijing, pp 189
Fougère-Danezan M, Joly S, Bruneau A, Gao XF, Zhang LB (2015) Phylogeny and biogeography of wild roses with specifc attention to polyploids. Annals Bot 115:275–291
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: Computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279
Gao FL, Chen CJ, Arab DA, Du ZG, He YH, Simon YW (2019) EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol Evol 9:3891–3898
Gonalves DJP, Jansen RK, Ruhlman TA (2020) Under the rug: Abandoning persistent misconceptions that obfuscate organelle evolution. Mol Phylogenet Evol 151:106903
Greiner S, Rauwolf U, Meurer J, Herrmann RG (2011) The role of plastids in plant speciation. Mol Ecol 20:671–691
Gu CZ (2003) Flora of China, vol 9. Science Press, Beijing, pp 360–445
Gudin S (2000) Rose: genetics and breeding. Plant Breed Rev 17:159–189
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Mol Phylogenet Evol 111:76–86
Huang DI, Cronk QC (2015) Plann: A command-line application for annotating chloroplast genome sequences. Appl Plant Sci 3:1500026
Jeon JH, Kim SC (2019) Comparative analysis of the complete chloroplast genome sequences of three closely related East-Asian wild Roses (Rosa sect. Synstylae; Rosaceae). Genes 10:23
Jian YH, Zhang YH, Yan HJ, Qiu XQ, Wang QG, Li SB, Zhang SD (2018) The complete chloroplast genome of a key ancestor of modern roses, Rosa chinensis var. spontanea, and a comparison with congeneric species. Molecules 23:389
Joly S, Bruneau A (2007) Delimiting species boundaries in Rosa Sect. Cinnamomeae (Rosaceae) in Eastern North America. Syst Bot 32:819–836
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kawamura K, Hibrand-Saint Oyant L, Crespel L, Thouroude T, Lalanne D, Foucher F (2011) Quantitative trait loci for flowering time and inflorescence architecture in rose. Theor Appl Genet 122:661–675
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kikuchi S, Bedard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339:571–574
Kim KJ, Lee HL (2004) Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res 11:247–261
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Koopman WJM, Wissemann V, Cock DK, van Huylenbroeck J, de Riek J, Sabatlno GJH, Visser D, Vosman B, Ritz CM, Maes B, Werlemark G, Nybom H, Debener T, Linde M, Smulders MJM (2008) AFLP markers as a tool to reconstruct complex relationships: a case study in Rosa (Rosaceae). Am J Bot 95:353–366
Ku TC, Robertson KR (2003) Rosa (Rosaceae). Flora of China 9, Science Press, Beijing & Missouri Botanical Garden Press, St. Louis, pp 339–381
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34:772−773
Lewis WH, Ertter B, Bruneau A (2015) Flora of North America 9, Family Rosaceae. Oxford University Press, London, pp 75–119
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lopes AS, Pacheco TG, Santos KGD, Vieira LN, Guerra MP, Nodari RO, de Souza EM, Pedrosa FO, Rogalski M (2018) The Linum usitatissimum L. plastome reveals atypical structural evolution, new editing sites, and the phylogenetic position of Linaceae within Malpighiales. Plant Cell Rep 37:307–328
Lowe TM, Chan PP (2016) tRNAscan-SE on-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57
Löytynoja A, Goldman N (2008) Phylogeny aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320:1632–1635
McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter DL, Hawksworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, Proud J, Proud’ Homme van Reine WF, Smith JF, Wiersema JH (2012) International code of nomenclature for algae, fungi and plants (Melbourne code): adopted by the eighteenth International botanical congress, Melbourne, Australia, July 2011. Regnum Vegetable 154:1–274
Mower JP (2005) PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinformatics 6:96
Mower JP (2009) The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res 37:W253–W259
Nguyen L-T, Schmidt HA, Arndt VH, Quang MB (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Bio Evol 32:268–274
Peden JF (1999) Analysis of codon usage. University of Nottingham, UK
Qu XJ, Moore MJ, Li DZ, Yi TS (2019) PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15:1–12
Rehder A (1940) Manual of cultivated trees and shrubs hardy in North America, 2nd edn. Macmillan, New York
Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302
Schwarz EN, Ruhlman TA, Weng ML, Khiyami MA, Sabir JSM, Hajarah NH (2017) Plastome-wide nucleotide substitution rates reveal accelerated rates in Papilionoideae and correlations with genome features across legume subfamilies. J Mol Evol 84:187–203
Seringe NC (1818) Observations générales sur les Roses. In: Musée helvétiqued histoire naturelle, (part Botanique). tome I. Genève: Chez l’auteur, Berne, pp 2.
Sharp PM, Li WH (1987) The codon adaptation index – a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15:1281–1295
Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, Burns P, Davis TM, Slovin JP, Bassil N, Hellens RP, Evans C, Harkins T, Kodira C, Desany B, Crasta OR, Jensen RV, Allan AC, Michael TP, Setubal JC, Celton JM, Rees DJ, Williams KP, Holt SH, Ruiz Rojas JJ, Chatterjee M, Liu B, Silva H, Meisel L, Adato A, Filichkin SA, Troggio M, Viola R, Ashman TL, Wang H, Dharmawardhana P, Elser J, Raja R, Priest HD, Bryant DW Jr, Fox SE, Givan SA, Wilhelm LJ, Naithani S, Christoffels A, Salama DY, Carter J, Lopez Girona E, Zdepski A, Wang W, Kerstetter RA, Schwab W, Korban SS, Davik J, Monfort A, Denoyes-Rothan B, Arus P, Mittler R, Flinn B, Aharoni A, Bennetzen JL, Salzberg SL, Dickerman AW, Velasco R, Borodovsky M, Veilleux RE, Folta KM (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43:109–116
Sudhir K, Glen S, Koichiro T (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Sun ZQ, Shi S, Pang XC, Chen JF, Wu YB (2020) Characterization of the complete chloroplast genome of Rosa laevigata var. leiocarpus in China and phylogenetic relationships. Mitochondrial DNA B 5:1996−1997
Thompson JD, Higgins DG (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thory CA (1820) Prodrome de la monographie des espèces et variétés connues du genre rosier, divisées selon leur ordre naturel. P. Dufart, Paris, pp 128
Vaezi J, Arjmandi AA, Sharghi HR (2019) Origin of Rosa ×binaloudensis (Rosaceae), a new natural hybrid species from Iran. Phytotaxa 411:23–28
Volker K (2011) When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci 68:567–586
Wang MC, Zhang C, Li MM, Gao XF (2019) The complete chloroplast genome sequence of Rosa banksiae var. normalis (Rosaceae). Mitochondrial DNA B 4:969–970
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. Academic Press Inc, London, pp 315–322
Wicke S, Schneeweiss GM, de Pamphilis CW, Muller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76:273–297
Wissemann V (2017) Conventional taxonomy (Wild Roses). Reference Module in Life Sciences, pp 1–6
Wissemann V, Ritz CM (2005) The genus Rosa (Rosoideae, Rosaceae) revisited: molecular analysis of nrITS-1 and atpB-rbcL intergenic spacer (IGS) versus conventional taxonomy. Bot J Linn Soc 147:275–290
Wu S, Ueda Y, Nishihara S, Matsumoto S (2001) Phylogenetic analysis of Japanese Rosa species using DNA sequences of nuclear ribosomal internal transcribed spacers (ITS). J Hortic Sci Biotech 76:127–132
Wu SQ, Ueda Y, He HY, Nishihara S, Matsumoto S (2005) Phylogenetic analysis of Japanese Rosa species using matK sequences. Breeding Sci 50:275–281
Xia X, Xie Z (2001) DAMBE: Software package for data analysis in molecular biology and evolution. J Hered 92:371–373
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang C, Ma YJ, Cheng BX, Zhou LJ, Yu C, Luo L, Pan HT, Zhang QX (2020) Molecular evidence for hybrid origin and phenotypic variation of Rosa section Chinenses. Genes 11:996
Yin XM, Liao BS, Guo S, Liang CL, Pei J, Xu J, Chen SL (2020) The chloroplasts genomic analyses of Rosa laevigata, R rugosa and R canina. Chin Med-UK 15:18
Zhang C, Xiong XH, Gao XF (2019) The complete chloroplast genome sequence of Rosa laevigata (Rosaceae). Mitochondrial DNA B 4:3556–3557
Zhang C, Li SQ, Zhang Y, Liu JQ, Gao XF (2020) Molecular and morphological evidence for hybrid origin and matroclinal inheritance of an endangered wild rose, Rosa×pseudobanksiae (Rosaceae) from China. Conserv Genet 21:1–11
Zhao YH, Lu DX, Han RB, Wang L, Qin P (2018) The complete chloroplast genome sequence of the shrubby cinquefoil Dasiphora fruticosa (Rosales Rosaceae). Conserv Genet Resour 10:675−678
Zhao L, Zhang H, Wang QG, Ma CL, Jian HY (2019) The complete chloroplast genome of Rosa lucidissima a critically endangered wild rose endemic to China. Mitochondrial DNA B 4:1826−41827
Zhu ZM, Gao XF, Fougere-Danezan M (2015) Phylogeny of Rosa sections Chinenses and Synstylae (Rosaceae) based on chloroplast and nuclear markers. Mol Phylogenet Evol 87:50–64
Acknowledgements
We sincerely thank Mr. Zheng-Zhi Jiang and Sao-Zong Yang for their collection of samples. The research was partially supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK0502), the National Natural Science Foundation of China (Grant No. 31670192) and the Science and Technology Basic Work (Grant No. 2017FY100104) to GXF.
Author information
Authors and Affiliations
Contributions
GXF offered study supervision, revised this manuscript draft and acquired funds. ZC conceived and designed the experiments and wrote the manuscript. LSQ, XHH, and LJQ helped to analyze the data. All the authors read and approved this manuscript for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by W.-W. Guo
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, C., Li, SQ., Xie, HH. et al. Comparative plastid genome analyses of Rosa: Insights into the phylogeny and gene divergence. Tree Genetics & Genomes 18, 20 (2022). https://doi.org/10.1007/s11295-022-01549-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-022-01549-8