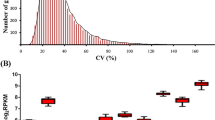
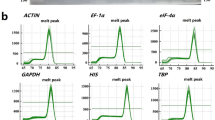
Abstract
Housekeeping genes were considered to have a constant expression without spatio-temporal difference. They are commonly used as the internal reference for PCR-based gene expression analysis. However, increasing evidence revealed that the expression of housekeeping genes also varied with tissues and treatments. Previous studies for reference gene identification were usually limited to a small number of housekeeping genes based on their expression stability comparison. In this study, reference genes for pear fruit were initially screened on the transcriptome level with 10 messenger RNA (mRNA)-seq datasets from the fruit of two pear species across important developmental stages. Ten potential reference genes were chosen for validation using the mRNA-seq data from fruit skin libraries. Five potential reference genes are proposed that can be used as suitable controls for PCR-based expression studies in pear fruit. Four of the selected genes had homologous annotation of important roles in the basic biological processes. Sulfhydryl oxidase 2 (SOX2) and protein phosphatase (PP) 2A also showed high stable expression in both pear fruit and other tissues and should be more widely applicable as reference genes in different tissues or species. This study provided a comprehensive view for transcriptional analysis in pear fruit, which will help researchers to select reference genes for normalization against target genes by qRT-PCR across different developmental stages of the fruit as well as for different tissues and species.





Similar content being viewed by others
References
Alejandro S, Rodríguez PL, Bellés JM, Yenush L, García-Sanchez MJ, Fernández JA, Serrano R (2007) An Arabidopsis quiescin-sulfhydryl oxidase regulates cation homeostasis at the root symplast-xylem interface. EMBO J 26:3203–3215
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–33402
Bowen J, Ireland HS, Crowhurst R, Luo Z, Watson A, Foster T, Gapper N, Giovanonni J, Mattheis J, Watkins C, Rudell D, Johnston J, Schaffer R (2014) Selection of low-variance expressed Malus × domestica (apple) genes for use as quantitative PCR reference genes (housekeepers). Tree Genet Genomes 10:751–759
Chen J, Li X, Wang D, Li L, Zhou H, Liu Z, Wu J, Wang P, Jiang X, Fabrice MR, Zhang S, Wu J (2015) Identification and testing of reference genes for gene expression analysis in pollen of Pyrus bretschneideri. Sci Hortic 190:43–56
Cheng X, Li M, Li D, Zhang J, Jin Q, Sheng L, Cai Y, Lin Y (2017) Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol Open 6:1602–1613
Chung S, Zhou Z, Huddleston KA, Harrison DA, Reed R, Coleman TA, Rymond BC (2002) Crooked neck is a component of the human spliceosome and implicated in the splicing process. Biochim Biophys Acta 1576:287–297
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Granados JM, Ávila C, Cánovas FM, Cañas RA (2016) Selection and testing of reference genes for accurate RT-qPCR in adult needles and seedlings of maritime pine. Tree Genet Genomes 12:60
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription–polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618
Imai T, Ubi BE, Saito T, Moriguchi T (2014) Evaluation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in Pyrus pyrifolia using different tissue samples and seasonal conditions. PLoS One 9:e86492
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Lefever S, Hellemans J, Pattyn F, Przybylski DR, Tayor C, Geurts R, Untergasser A, Vandesompele J (2009) RDML: structured language and reporting guide-lines for real-time quantitative PCR data. Nucleic Acids Res 37:2065–2069
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Saraiva KD, Fernandes de Melo D, Morais VD, Vasconcelos IM, Costa JH (2014) Selection of suitable soybean EF1α genes as internal controls for real-time PCR analyses of tissues during plant development and under stress conditions. Plant Cell Rep 33:1453–1465
Wang Z, Gerstein M, Snyder M (2009) RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang Y, Dai M, Zhang S, Shi Z (2014a) Exploring candidate genes for pericarp russet pigmentation of sand pear (Pyrus pyrifolia) via RNA-seq data in two genotypes contrasting for pericarp color. PLoS One 9:e83675
Wang Y, Zhang S, Dai M, Shi Z (2014b) Pigmentation in sand pear (Pyrus pyrifolia) fruit: biochemical characterization, gene discovery and expression analysis with exocarp pigmentation mutant. Plant Mol Biol 85:123–134
Wang L, Chen Y, Wang S, Xue H, Su Y, Yang J, Li X (2017) Identification of candidate genes involved in the sugar metabolism and accumulation during pear fruit post-harvest ripening of ‘Red Clapp’s Favorite’ (Pyrus communis L.) by transcriptome analysis. Hereditas. 155:11
Wang Y, Dai M, Cai D, Miao L, Wei L, Shi Z (2018) Characterizing the expression of translation elongation factor gene EF1α in pear (Pyrus) fruit: evaluation of EF1α as a housekeeping gene. Tree Gene Genomes 14:62
Wang Y, Dai M, Cai D, Shi Z (2019) Expression stability analysis of common internal reference genes in pear fruit based on high-throughput sequencing. Mol Plant Breed 17:746–753 (in Chinese)
Wu T, Zhang R, Gu C, Wu J, Wan H, Zhang S, Zhang S (2012) Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Afr J Agric Res 7:3701–3704
Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, Chen NJ, Nishio T, Xu X, Cong L, Qi K, Huang X, Wang Y, Zhao X, Wu J, Deng C, Gou C, Zhou W, Yin H, Qin G, Sha Y, Tao Y, Chen H, Yang Y, Song Y, Zhan D, Wang J, Li L, Dai M, Gu C, Wang Y, Shi D, Wang X, Zhang H, Zeng L, Zheng D, Wang C, Chen M, Wang G, Xie L, Sovero V, Sha S, Huang W, Zhang S, Zhang M, Sun J, Xu L, Li Y, Liu X, Li Q, Shen J, Wang J, Paull RE, Bennetzen JL, Wang J, Zhang S (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Xu Y, Li H, Li X, Lin J, Wang Z, Yang Q, Chang Y (2015) Systematic selection and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear. Acta Physiol Plant 37:40
Yang YN, Yao GF, Zheng D, Zhang SL, Wang C, Zhang MY, Wu J (2015) Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep 34:189–198
Data archiving statement
Pear gene sequences and annotation information presented in this report are available in the pear genome database (Pear Genome Project, http://peargenome.njau.edu.cn/) and NCBI database (https://www.ncbi.nlm.nih.gov/). The apple genes are available in Malus all species | GDR (https://www.rosaceae.org/species/malus/all).
Funding
This research was partially supported by the Special Funds for China Agriculture Research System (CARS-28-03); the Key Project for New Agricultural Cultivar Breeding in Zhejiang Province, China (2016C02052); and Zhejiang Provincial Natural Science Foundation of China (LY15C150003).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Chagné
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplemental Figure S1
Quality detection of different total RNA samples by agarose gel electrophoresis. CK1, water control. CK2, total RNA control. S1~S9, different total RNA samples for the qRT-PCR analysis. (PDF 30 kb)
Supplemental Figure S2
The quality testing results of part of total RNA samples by Agilent 2100 bioanalyzer. (PDF 358 kb)
Supplemental Figure S3
Ct values of the six candidate reference genes tested in the eight leaf samples of different pear genotypes. (A) Ct values of the six candidate reference genes with three replicates. (B) The mean Ct values of the six candidate reference genes in all pear samples. The bars indicate the maximum and minimum Ct values. (PDF 52 kb)
Supplemental Table S1
Analysis of the expression stability of Actin genes using the RNA-seq data in ten different samples of pear fruit. (XLSX 12 kb)
Supplemental Table S2
Expression stability of the candidate reference genes in leaf tissues of the eight pear genotypes calculated by BestKeeper and coefficient of variation. (DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Dai, M., Cai, D. et al. Screening for quantitative real-time PCR reference genes with high stable expression using the mRNA-sequencing data for pear. Tree Genetics & Genomes 15, 54 (2019). https://doi.org/10.1007/s11295-019-1361-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-019-1361-6