Abstract
European hazelnut (Corylus avellana L.) is an economically and nutritionally important nut crop with wild and cultivated populations found throughout Europe and in parts of Asia. This study examined the molecular genetic diversity and population structure of 402 genotypes including 143 wild individuals, 239 landraces, and 20 cultivars from the Turkish national hazelnut collection using simple sequence repeat (SSR) markers. A total of 30 SSR markers yielded 407 polymorphic fragments. Diversity analysis of the Turkish hazelnut genotypes indicated that they fell into three subpopulations according to ad hoc statistics and neighbor-joining algorithm. Although all cultivars clustered together, they overlapped with the wild accessions and landraces. Thus, the dendrogram, principal coordinate, and population structure analyses suggest that they share the same gene pool. A total of 78 accessions were selected as a core set to encompass the molecular genetic and morphological diversity present in the national collection. This core set should have priority in preservation efforts and in trait characterization.





Similar content being viewed by others
References
Alasalvar C, Shahidi F, Liyanapathirana CM, Ohshima T (2003) Turkish tombul hazelnut (Corylus avellana L.). 1. Compositional characteristics. J Agric Food Chem 51:3790–3796
Bacchetta L, Avanzato D, Di Giovanni B, Botta R, Boccacci P, Drogoudi P, Metzidakis I, Rovira M, Sarraquigne JP, Silva AP, Solar A, Spera D (2014) The reorganisation of European hazelnut genetic resources in the SAFENUT (AGRI GEN RES) project. Acta Hort 1052:67–74
Balik HI, Balik SK, Okay AN (2015) Yeni Fındık Çeşitleri (Okay 28 ve Giresun Melezi). Harran Tarım ve Gıda Bilimleri Dergisi 19:104–109
Balik HI, Balik SK, Beyhan N, Erdogan V (2016) Hazelnut cultivars, ISBN 978-605-137-559-5
Bassil NV, Botta R, Mehlenbacher SA (2005) Additional microsatellite markers of the European hazelnut. Acta Hort 686:105
Bassil N, Boccacci P, Botta R, Postman J, Mehlenbacher SA (2013) Nuclear and chloroplast microsatellite markers to assess genetic diversity and evolution in hazelnut species, hybrids and cultivars. Gen Res Crop Evol 60:543–568
Beltramo C, Valentini N, Portis E, Marinoni DT, Boccacci P, Prando MAS, Botta R (2016) Genetic mapping and QTL analysis in European hazelnut (Corylus avellana L.) Mol Breed 36:1–17
Boccacci P, Botta R (2010) Microsatellite variability and genetic structure in hazelnut (Corylus avellana L.) cultivars from different growing regions. Sci Hortic 124:128–133
Boccacci P, Akkak A, Bassil NV, Mehlenbacher SA, Botta R (2005) Characterization and evaluation of microsatellite loci in European hazelnut (Corylus avellana L.) and their transferability to other Corylus species. Mol Ecol Notes 5:934–937
Boccacci P, Akkak A, Botta R (2006) DNA typing and genetic relationship among European hazelnut (Corylus avellana L.) cultivars using microsatellite markers. Genome 49:598–611
Boccacci P, Botta R, Rovira M (2008) Genetic diversity of hazelnut (Corylus avellana L.) germplasm in northeastern Spain. Hort Sci 43:667–672
Boccacci P, Aramini M, Valentini N, Bacchetta L, Rovira M, Drogoudi P, Silva AP, Solar A, Calizzano F, Erdogan V, Cristoferi V, Ciarmiello LF, Contessa C, Ferreira JJ, Marra FP, Botta R (2013) Molecular and morphological diversity of on-farm hazelnut (Corylus avellana L.) landraces from southern Europe and their role in the origin and diffusion of cultivated germplasm. Tree Genet. Genomes 9:1465–1480
Boccacci P, Beltramo C, Prando MS, Lembo A, Sartor C, Mehlenbacher SA, Botta R, Marinoni DT (2015) In silico mining, characterization and cross-species transferability of EST-SSR markers for European hazelnut (Corylus avellana L.) Mol Breed 35:1–14
Brown AHD (1995) The core collection at the crossroads. In: Hodgkin T, Brown AHD, van Hintum TJL, Morales EAV (eds) Core collections of plant genetic resources. Wiley, London, pp 3–19
Caliskan T, Cetiner E (1997) Characterization studies on some hazelnut cultivars and types. In IV International Symposium on Hazelnut 445:2–12
Campa A, Trabanco N, Pérez-Vega E, Rovira M, Ferreira JJ (2011) Genetic relationship between cultivated and wild hazelnuts (Corylus avellana L.) collected in northern Spain. Plant Breed 130:360–366
Colburn BC, Mehlenbacher SA, Sathuvalli VR (2017) Development and mapping of microsatellite markers from transcriptome sequences of European hazelnut (Corylus avellana L.) and use for germplasm characterization. Mol Breed 37:16
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Food and Agriculture Organization of the United Nations, FAOSTAT (2013) http://www.fao.org/faostat/en/#data/TP Accessed 10.01.2017
Food and Agriculture Organization of the United Nations, FAOSTAT (2014) http://www.fao.org/faostat/en/#data/QC Accessed 10.01.2017
Frankel OH (1984) Genetic perspectives of germplasm conservation. In: Arber W, Llimensee K, Peacock WJ, Starlinger P (eds) Genetic manipulation: impact on man and society. Cambridge University Press, Cambridge, pp 161–170
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–207
Gao H, Williamson S, Bustamante CD (2007) A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176:1635–1651
Gökirmak T, Mehlenbacher SA, Bassil NV (2009) Characterization of European hazelnut (Corylus avellana L.) cultivars using SSR markers. Genet Resour Crop Evol 56:147–172
Gurcan K, Mehlenbacher SA (2010) Development of microsatellite marker loci for European hazelnut (Corylus avellana L.) from ISSR fragments. Mol Breed 26:551–559
Gurcan K, Mehlenbacher SA, Botta R, Boccacci P (2010a) Development, characterization, segregation, and mapping of microsatellite markers for European hazelnut (Corylus avellana L.) from enriched genomic libraries and usefulness in genetic diversity studies. Tree Genet Genomes 6:513–531
Gurcan K, Mehlenbacher SA, Erdogan V (2010b) Genetic diversity in hazelnut (Corylus avellana L.) cultivars from Black Sea countries assessed using SSR markers. Plant Breed 129:422–434
Ives C, Sathuvalli VR, Colburn BC, Mehlenbacher SA (2014) Mapping the incompatibility and style color loci in two hazelnut progenies. Hortscience 49:250–253
Kim KW, Chung HK, Cho GT, Ma KH, Chandrabalan D, Gwag JG, Kim TS, Cho EG, Park YJ (2007) PowerCore: a program applying the advanced M strategy with a heuristic search for establishing allele mining sets. Bioinformatics 23:2155–2162
Koksal, AI (2002) Turkish hazelnut cultivars. Hazelnut Promotion Group, Ankara ISBN, 975–92886
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Martins S, Simões F, Mendonça D, Matos J, Silva AP, Carnide V (2013) Chloroplast SSR genetic diversity indicates a refuge for Corylus avellana in northern Portugal. Genet Resour Crop Evol 60:1289–1295
Martins S, Simões F, Matos J, Silva AP, Carnide V (2014) Genetic relationship among wild, landraces and cultivars of hazelnut (Corylus avellana L.) from Portugal revealed through ISSR and AFLP markers. Plant Syst Evol 300:1035–1046
Martins S, Simões F, Mendonça D, Matos J, Silva AP, Carnide V (2015) Western European wild and landraces hazelnuts evaluated by SSR markers. Plant Mol Biol Rep: 1–9
Mehlenbacher SA, Brown RN, Nouhra ER, Gokirmak T, Bassil NV, Kubisiak TL (2006) A genetic linkage map for hazelnut (Corylus avellana L.) based on RAPD and SSR markers. Genome 49:122–133
Ozturk SC, Ozturk SE, Celik I, Stampar F, Veberic R, Doganlar S, Solar A, Frary A (2017) Molecular genetic diversity and association mapping of nut and kernel traits in Slovenian hazelnut (Corylus avellana) germplasm. Tree Genet Genomes 13:16
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. Available: http://darwin.cirad.fr/darwin
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000) Association mapping in structured populations. Am J Hum Genet 67:170–181
Sathuvalli VR, Chen H, Mehlenbacher SA, Smith DC (2011) DNA markers linked to eastern filbert blight resistance in “Ratoli” hazelnut (Corylus avellana L.). Tree Genet. & Genomes 7:337-345
Valentini N, Calizzano F, Boccacci P, Botta R (2014) Investigation on clonal variants within the hazelnut (Corylus avellana L.) cultivar ‘Tonda Gentile delle Langhe’. Sci Hortic 165:303–310
Acknowledgments
This study was supported by The Scientific and Technological Research Council of Turkey (TUBITAK, project no: 212T201).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data archiving statement
Data will be available at http://plantmolgen.iyte.edu.tr/data/ upon publication.
Additional information
Communicated by Z. Kaya
Electronic supplementary material
Suppl. Table 1
(DOCX 47 kb)
Suppl. Figure 1
Map of Turkey’s Black Sea region where hazelnut accessions were collected. Light blue star: Duzce; dark green star: Kastamonu; yellow star: Sinop; green star: Samsun; blue star: Ordu; black star: Giresun; red star: cultivars; fuchsia star: Trabzon; brown star: Rize and gray star: Artvin. Erzurum (not shown) is located in eastern Anatolia region and south of Rize and Artvin. Red arrows show the area expanded in the lower map with yellow stars indicating original collection locations of accessions. (GIF 282 bytes)
Suppl. Figure 2
Delta K values of the Structure program outcome for each subpopulation assumption. The value of K with the highest Delta K value was chosen as the best number of subpopulations for the hazelnut accessions (K = 2). (GIF 541 bytes)
Suppl. Figure 3
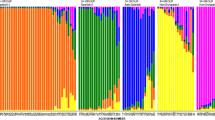
Population structure of Turkish hazelnuts based on SSR results. Each accession is represented by a vertical bar. Green sections within each vertical bar indicate membership coefficient (y-axis) of the accession to subpopulation 1 while red sections indicate membership to subpopulation 2. (GIF 10 kb)
Rights and permissions
About this article
Cite this article
Öztürk, S.C., Balık, H.İ., Balık, S.K. et al. Molecular genetic diversity of the Turkish national hazelnut collection and selection of a core set. Tree Genetics & Genomes 13, 113 (2017). https://doi.org/10.1007/s11295-017-1195-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1195-z