Abstract
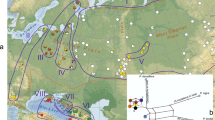
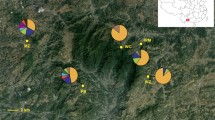
Genetic differentiation of scattered populations at neutral loci is characterized by genetic drift counteracted by the remaining gene flow. Populations of Pinus cembra in the Carpathian Mountains are isolated and restricted to island-like stands at high-elevation mountain ranges. In contrast, paleobotanical data suggest an extended early Holocene distribution of P. cembra in the Carpathians and its surrounding areas, which has contracted to the currently disjunct occurrences. We analyzed the genetic variation of 11 Carpathian populations of P. cembra at chloroplast and, in part newly developed, nuclear microsatellites. Both marker types revealed low levels of genetic differentiation and a lack of isolation by distance, reflecting the post-glacial retraction of the species to its current distribution. Stronger effects of genetic drift were implied by the higher genetic differentiation found for haploid chloroplast than for diploid nuclear markers. Moreover, we found no association between the values of population genetic differentiation for the two marker types. Several populations indicated recent genetic bottlenecks and inbreeding as a consequence of decline in population sizes. Moreover, we found individuals in two populations from the Rodnei Mountains that strikingly differed in assignment probabilities from the remaining specimens, suggesting that they had been introduced from a provenance outside the studied populations. Comparison with Eastern Alpine P. cembra and individuals of the closely related Pinus sibirica suggests that these individuals presumably are P. sibirica. Our study highlights the importance of the maintenance of sufficiently large local population sizes for conservation due to low connectivity between local occurrences.





Similar content being viewed by others
References
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Blackwell, Oxford
Alsos IG, Alm T, Normand S, Brochmann C (2009) Past and future range shifts and loss of diversity in dwarf willow (Salix herbacea L.) inferred from genetics, fossils and modeling. Glob Ecol Biogeogr 18:223–239
Belokon MM, Belokon YS, Politov DV, Altukhov YP (2005) Allozyme polymorphism of Swiss stone pine Pinus cembra L. in mountain populations of the Alps and the Eastern Carpathians. Russ J Genet 41:1268–1280
Birky CW Jr, Fuerst P, Maruyama T (1989) Organelle gene diversity under migration, mutation, and drift: equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparison to nuclear genes. Genetics 121:613–627
Blada I (1997) Stone pine (Pinus cembra L.) provenance experiment in Romania. Silvae Genet 46:197–200
Blada I (2008) Pinus cembra distribution in the Romanian Carpathians. Ann For Res 51:115–132
Casalegno S, Amatulli G, Camia A, Nelson A, Pekkarinen A (2010) Vulnerability of Pinus cembra L. in the Alps and the Carpathian mountains under present and future climates. For Ecol Manag 259:750–761
Corander J, Tang J (2007) Bayesian analysis of population structure based on linked molecular information. Math Biosci 205:19–31
Corander J, Waldmann P, Sillanpää MJ (2003) Bayesian analysis of genetic differentiation between populations. Genetics 163:367–374
Critchfield WB (1986) Hybridization and classification of the white pines (Pinus section Strobus). Taxon 35:647–656
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annu Rev Ecol Syst 24:217–242
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Fekete L (1887) Abauj-Torna-, Szepes- és Gömör vármegyék erdő- tenyésztési viszonyai. Erdészeti lapok 26:525–549 (Silvicultural characteristics of Abauj-Torna, Szepes and Gömör counties)
Feurdean A, Tantau I, Farcas S (2011) Holocene variability in the range distribution and abundance of Pinus, Picea abies, and Quercus in Romania; implications for their current status. Quat Sci Rev 30:3060–3075
Goncharenko GG, Padutov VE, Silin AE (1992) Population structure, gene diversity, and differentiation in natural populations of Cedar pines (Pinus subsect. Cembrae, Pinaceae) in the USSR. Plant Syst Evol 182:121–134
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J (2002) Fstat 2.9.3.2. URL: http://www2.unil.ch/popgen/softwares/fstat.htm
Gugerli F, Senn J, Anzidei M, Madaghiele A, Büchler U, Sperisen C, Vendramin GG (2001) Chloroplast microsatellites and mitochondrial nad1 intron 2 sequences indicate congruent phylogenetic relationships among Swiss stone pine (Pinus cembra), Siberian stone pine (Pinus sibirica), and Siberian dwarf pine (Pinus pumila). Mol Ecol 10:1489–1497
Höhn M, Ábrán P, Vendramin GG (2005) Genetic analysis of Swiss stone pine populations (Pinus cembra L. subsp. cembra) from the Carpathians using chloroplast microsatellites. Acta Silv Lign Hung 1:39–47
Höhn M, Gugerli F, Ábrán P, Bisztray G, Buonamici A, Cseke K, Hufnagel L, Quintela-Sabarís C, Sebastiani F, Vendramin GG (2009) Variation in the chloroplast DNA of Swiss stone pine (Pinus cembra L.) reflects contrasting post-glacial history of populations from the Carpathians and the Alps. J Biogeogr 36:1798–1806
Höhn M, Hufnagel L, Cseke K, Vendramin GG (2010) Current range characteristics of Swiss stone pine (Pinus cembra L.) along the Carpathians revealed by chloroplast SSR markers. Acta Biol Hung 61(Suppl):61–67
Hu X-S, Ennos RA (1997) On estimation of the ratio of pollen to seed flow among plant populations. Heredity 79:541–552
Hu X-S, Ennos RA (1999) Impacts of seed and pollen flow on population genetic structure for plant genomes with three contrasting modes of inheritance. Genetics 152:441–450
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jankovská V (1984) Late glacial finds of Pinus cembra L. in the Lubovnianská kotlina Basin. Folia Geobot Phytotaxon 19:323–325
Jankovská V, Pokorný P (2008) Forest vegetation of the last full-glacial period in the Western Carpathians (Slovakia and Czech Republic). Preslia 80:307–324
Karhu A, Vogl C, Moran GF, Bell JC, Savolainen O (2006) Analysis of microsatellite variation in Pinus radiata reveals effects of genetic drift but no recent bottlenecks. J Evol Biol 19:167–175
Kimura M, Weiss GH (1964) The stepping stone model of population structure and the decrease of the genetic correlation with distance. Genetics 49:561–576
Krutovsky KV, Politov DV, Altukhov YP (1992) Genetic differentiation and phylogeny of stone pine species based on isozyme loci. International Workshop on Subalpine Stone Pines and Their Environment: The Status of Our Knowledge, St. Moritz, Switzerland, September 5-11, 1992, Proceedings 19–30
Kuneš P, Pelánková B, Chytrý M, Jankovská V, Pokorný P, Petr L (2008) Interpretation of the last-glacial vegetation of eastern-central Europe using modern analogues from southern Siberia. J Biogeogr 35:2223–2236
Latta RG, Mitton JB (1997) A comparison of population differentiation across four classes of gene marker in limber pine (Pinus flexilis James). Genetics 146:1153–1163
Leberg PL (2002) Estimating allelic richness: effects of sample size and bottlenecks. Mol Ecol 11:2445–2449
Lenormand T (2002) Gene flow and the limits to natural selection. Trends Ecol Evol 17:183–189
Magyari E, Jakab G, Bálint M, Kern Z, Buczkó K, Braun M (2012) Rapid vegetation response to Lateglacial and early Holocene climatic fluctuation in the South Carpathian Mountains (Romania). Quat Sci Rev 35:116–130
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Mattes H (1982) Die Lebensgemeinschaft von Tannenhäher und Arve, 2nd edn. Eidgenössische Forschungsanstalt für das forstliche Versuchswesen, Birmensdorf
McKay JK, Christian CE, Harrison S, Rice KJ (2005) “How local is local?”—A review of practical and conceptual issues in the genetics of restoration. Restor Ecol 13:432–440
Meusel H, Jäger E, Weinert E (1965) Vergleichende Chorologie der Zentraleuropäischen Flora. Gustav Fischer, Jena
Myers ER, Chung MY, Chung MG (2007) Genetic diversity and spatial genetic structure of Pinus strobus (Pinaceae) across an island landscape inferred from allozyme and cpDNA markers. Plant Syst Evol 264:15–30
Paryski W (1971) Sadzenie i przesadzanie limby (The planting and transplanting of Pinus cembra). In: Białobok S (ed) Limba Pinus cembra L. (The stone pine Pinus cembra L.) Nasze drzewa leśne 2:50–56 (in Polish with an English summary) cit. in: Zwijacz-Kozica T, Żywiec M (2007) Fifty-year changes in a strictly protected stone pine population in the Tatra National Park. Nat Conserv 64:73–82
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Petit RJ, Kremer A, Wagner DB (1993) Finite island model for organelle and nuclear genes in plants. Heredity 71:630–641
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701
Piry S, Luikart G, Cornuet J-M (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Provan J, Soranzo N, Wilson NJ, Goldstein DB, Powell W (1999) A low mutation rate for chloroplast microsatellites. Genetics 153:943–947
Provan J, Beatty GE, Hunter AM, McDonald RA, McLaughlin E, Preston SJ, Wilson S (2008) Restricted gene flow in fragmented populations of a wind-pollinated tree. Conserv Genet 9:1521–1532
Puşcaş M, Choler P, Tribsch A, Gielly L, Rioux D, Gaudeul M, Taberlet P (2008) Post-glacial history of the dominant alpine sedge Carex curvula in the European Alpine System inferred from nuclear and chloroplast markers. Mol Ecol 17:2417–2429
Ribeiro MM, Mariette S, Vendramin GG, Szmidt AE, Plomion C, Kremer A (2002) Comparison of genetic diversity estimates within and among populations of maritime pine using chloroplast simple-sequence repeat and amplified fragment length polymorphism data. Mol Ecol 11:869–877
Ronikier M (2011) Biogeography of high-mountain plants in the Carpathians: an emerging phylogeographical perspective. Taxon 60:373–389
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rowland V (1882) A czirbolya-fenyő (Pinus cembra) elöjövétele- és tenyésztéséről a központi Kárpátokban. Erdészeti lapok 21:422–427 (On the discovery of Swiss stone pine (Pinus cembra) and its propagation in the Central Carpathians)
Rybníčková E, Rybníček K (2006) Pollen and macroscopic analyses of sediments from two lakes in the High Tatra mountains, Slovakia. Veget Hist Archeobot 15:345–356
Salzer K, Sebastiani F, Gugerli F, Bounamici A, Vendramin GG (2009) Isolation and characterization of polymorphic nuclear microsatellite loci in Pinus cembra L. Mol Ecol Resour 9:858–861
Schoebel CN, Brodbeck S, Buehler D, Cornejo C, Gajurel J, Hartikainen H, Keller D, Leys M, Říčanová Š, Segelbacher G, Werth S, Csencsics D (2013) Lessons learned from microsatellite development for nonmodel organisms using 454 pyrosequencing. J Evol Biol 26:600–611
Sherwin WB, Jabot F, Rush R, Rosetto M (2006) Measurement of biological information with applications from genes to landscapes. Mol Ecol 15:2857–2869
Szmidt AE (1982) Genetic variation in isolated populations of stone pine (Pinus cembra). Silva Fenn 16:196–200
Thiel-Egenter C, Holderegger R, Brodbeck S, Intrabiodiv Consortium, Gugerli F (2009) Concordant genetic breaks, identified by combining clustering and tessellation methods, in two co-distributed alpine plant species. Mol Ecol 18:4495–4507
Tomback DF, Holtmeier F-K, Mattes H, Carsey KS, Powell ML (1993) Tree clusters and growth form distribution in Pinus cembra, a bird-dispersed pine. Arct Alp Res 25:374–381
Vendramin GG, Lelli L, Rossi P, Morgante M (1996) A set of primers for the amplification of 20 chloroplast microsatellites in Pinaceae. Mol Ecol 5:595–598
Voronoï MG (1908) Nouvelles application des paramètres continus à la théorie des formes quadratiques. Deuxième mémoire Recherche sur le paralléloedres primitifs. J Reine Angew Math 134:198–207
Wachowiak W, Boratyńska K, Cavers S (2013) Geographical patterns of nucleotide diversity and population differentiation in three closely related European pine species in the Pinus mugo complex. Bot J Linn Soc 172:225–238
Willis KJ, Van Andel TH (2004) Trees or no trees? The environments of central and eastern Europe during the Last Glaciation. Quat Sci Rev 23:2369–2387
Willis KJ, Rudner E, Sümegi P (2000) The full-glacial forests of central and southeastern Europe. Quat Res 53:203–213
Wohlfarth B, Hannon G, Feurdean A, Ghergari L, Onac BP, Possnert G (2001) Reconstruction of climatic and environmental changes in NW Romania during the early part of the last deglaciation (~15,000–13,600 cal yr BP). Quat Sci Rev 20:1897–1914
Acknowledgments
We acknowledge the CRUS-Sciex grant (12.071) that allowed the stay of BL at WSL.
Data archiving statement
Sequence data of nuclear microsatellite loci are retrievable from NCBI GenBank; cp/nSSR data are uploaded on the TreeGenes database.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. G. Vendramin
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 592 kb)
Rights and permissions
About this article
Cite this article
Lendvay, B., Höhn, M., Brodbeck, S. et al. Genetic structure in Pinus cembra from the Carpathian Mountains inferred from nuclear and chloroplast microsatellites confirms post-glacial range contraction and identifies introduced individuals. Tree Genetics & Genomes 10, 1419–1433 (2014). https://doi.org/10.1007/s11295-014-0770-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0770-9