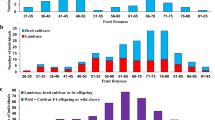
Abstract
Upon crushing, amygdalin present in bitter almonds is hydrolysed to benzaldehyde, which gives a bitter flavour, and to cyanide, which is toxic. Bitterness is attributable to the recessive allele of the Sweet kernel (Sk/sk) gene and is selected against in breeding programmes. Almond has a long intergeneration period due to its long juvenile phase, so breeders must wait 3 or 4 years to evaluate fruit traits in the field. For this reason, it is important to develop molecular markers to distinguish between sweet and bitter genotypes. The Sk gene is known to map to linkage group five (G5) of the almond genome, but its function is still undefined. Candidate genes involved in the amygdalin pathway have been mapped, but none of them were located to G5. We have saturated G5 with additional Simple Sequence Repeats (SSRs) using the progeny from the cross “R1000” × “Desmayo Largueta” and found six SSRs (UDA-045, EPDCU2584, CPDCT028, BPPCT037, PceGA025, and CPDCT016) closely linked to the Sk locus. The genotypes of four of these SSRs flanking the Sk locus, in a number of parents and a few seedlings of the CEBAS-CSIC almond breeding programme, allowed us to estimate the haplotypes of the parents, identifying the marker alleles adequate for an early and highly efficient selection against bitter genotypes. This analysis has established the usefulness of SSRs for screening populations of fruit trees such as almond by an easy, polymerase chain reaction-based method.




Similar content being viewed by others
References
Arús P, Vargas FJ, Romero M (1992) Linkage analysis of isozyme genes in almond. In: Grasselly C (ed) Eighth Coll GREMPA. Office des Communatés Européennes, Luxembourg, pp 201–207
Arús P, Yamamoto T, Dirlewanger E, Abbott AG (2005) Synteny in the Rosaceae. Plant Breed Rev 27:175–211
Ballester J, Socias I, Company R, Arús P, de Vicente MC (2001) Genetic mapping of a major gene delaying blooming time in almond. Plant Breed 120:268–270
Bonfield JK, Smith KF, Staden RA (1995) New DNA sequence assembly program. Nucleic Acids Res 23:4992–4999
Chandler WH (1957) In: Deciduous orchards. Lea and Febiger, Philadelphia
Dicenta F, García JE (1993) Inheritance of the kernel flavour in almond. Heredity 70:308–312
Dicenta F, Martínez-Gómez P, Ortega E (2000) Cultivar pollinizer does not affect almond flavor. HortScience 35:1156–1154
Dicenta F, Martínez-Gómez P, Grané N, Martín ML, León A, Cánovas JA, Berenguer V (2002) Relationship between cyanogenic compounds in kernels, leaves, and roots of sweet and bitter kernelled almonds. J Agric Food Chem 50:2149–2152
Dicenta F, Ortega E, Martinez-Gomez P (2007) Use of recessive homozygous genotypes to assess the genetic control of kernel bitterness in almond. Euphytica 153:221–225
Dirlewanger E, Crosson A, Tavaud P, Aranzana MJ, Poizat C, Zanetto A, Arús P, Laigret L (2002) Development of microsatellite markers in peach and their use in genetic diversity analysis in peach and sweet cherry. Theor Appl Genet 105:127–138
Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldré F, Cosson P, Howad W, Arús P (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci USA 101:9891–9896
Evreinoff VA (1952) Quelques observations biologiques sur l’amandier. Rev Int Bot Appl Agric Trop 32:442–459
Franks TK, Yadollahi A, Wirthensohn MG, Guerin JR, Kaiser BN, Sedgley M, Ford CM (2008) A seed coat cyanohydrin glucosyltransferase is associated with bitterness in almond (Prunus dulcis) kernels. Funct Plant Biol 35:236–246
Heppner J (1923) The factor for bitterness in the sweet almond. Genetics 8:390–392
Heppner J (1926) Further evidence on the factor for bitterness in the sweet almond. Genetics 11:605–606
Howad W, Yamamoto T, Dirlewanger E, Testolin R, Cosson P, Cipriani G, Monforte AJ, Georgi L, Abbott AG, Arús P (2005) Mapping with a few plants: using selective mapping for microsatellite saturation of the Prunus reference map. Genetics 171:1305–1309
Joobeur T (1998) Construcción de un mapa de marcadores moleculares y análisis genético de caracteres agronómicos en Prunus. PhD Thesis. Universitat de Lleida, Lleida, Spain
Joobeur T, Viruel MA, de Vicente MC, Jáuregui B, Ballester J, Dettori MT, Verde I, Truco MJ, Messeguer R, Battle I, Quarta R, Dirlewanger E, Arús P (1998) Construction of a saturated linkage map for Prunus using an almond × peach F2 progeny. Theor Appl Genet 97:1034–1041
Kester DE, Asay RN (1975) Almonds. In: Janick J, Moore JN (eds) Advances in fruit breeding. Purdue University Press, Indiana
Lander ES, Green P, Abrahamson J, Barlow AM, Daly MJ, Lincoln SE, Newbur L (1987) MapMaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
McCarty CD, Leslie JW, Frost HB (1952) Bitterness of kernels of almond x peach hybrids and their parents. Proc Am Soc Hortic Sci 59:254–258
Mnejja M, Garcia-Mas J, Howad W, Arús P (2005) Development and transportability across Prunus species of 42 polymorphic almond microsatellites. Mol Ecol Notes 5:531–535
Negri P, Bassi D, Magnanini E, Rizzo M, Bartolozzi F (2008) Bitterness inheritance in apricot (P. armeniaca L) seeds. Tree Genetic Genomes 4:767–776
Sánchez-Pérez R, Howad W, Dicenta F, Arús P, Martínez-Gómez P (2007) Mapping major genes and quantitative trait loci controlling agronomic traits in almond. Plant Breeding 126:310–318
Sánchez-Pérez R, Jørgensen K, Olsen CE, Dicenta F, Møller BL (2008) Bitterness in almonds. Plant Physiol 146:1040–1052
Testolin R, Messina R, Lain O, Marrazo T, Huang G, Cipriani G (2004) Microsatellites isolated in almond from an AC-repeat enriched library. Mol Ecol Notes 4:459–461
Vargas FJ, Romero MA, Batlle I (2001) Kernel taste inheritance in almond. Options Méditerr 56:129–134
Werner DJ, Creller MA (1997) Genetic studies in peach: inheritance of sweet kernel and male sterility. J Am Soc Hortic Sci 122:215–217
Woodroof JG (1967) Tree nuts: production, processing, products, vol I. AVI Publishing, New York
Zhou J, Hartmann S, Shepherd BK, Poulton JE (2002) Investigation of the microheterogeneity and aglycone specificity-conferring residues of black cherry prunasin hydrolases. Plant Phys 129:1252–1264
Acknowledgements
This study was financed by a project of the Spanish Ministerio de Educación y Ciencia. We wish to thank F. Rook, A. Takos, V. Fuller, R. Lopez, and C. Martin for help in writing the manuscript and T. Cremades and M. Gambín for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table A
Pedigree and genotypes of the bitterness (Sk/sk) gene and four tightly linked SSRs of almond cultivars (from “Del Cid” to “Tardy Nonpareil”) and progeny (seedlings with a capital letter followed by a number) from the CEBAS-CSIC almond breeding programme. Markers and the Sk/sk gene are ordered according to their map position. The marker allele linked to the sk allele is in bold (PDF 58 kb)
Rights and permissions
About this article
Cite this article
Sánchez-Pérez, R., Howad, W., Garcia-Mas, J. et al. Molecular markers for kernel bitterness in almond. Tree Genetics & Genomes 6, 237–245 (2010). https://doi.org/10.1007/s11295-009-0244-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-009-0244-7