Abstract
Automating ontology construction and curation is an important but challenging task in knowledge engineering and artificial intelligence. Prediction by machine learning techniques such as contextual semantic embedding is a promising direction, but the relevant research is still preliminary especially for expressive ontologies in Web Ontology Language (OWL). In this paper, we present a new subsumption prediction method named BERTSubs for classes of OWL ontology. It exploits the pre-trained language model BERT to compute contextual embeddings of a class, where customized templates are proposed to incorporate the class context (e.g., neighbouring classes) and the logical existential restriction. BERTSubs is able to predict multiple kinds of subsumers including named classes from the same ontology or another ontology, and existential restrictions from the same ontology. Extensive evaluation on five real-world ontologies for three different subsumption tasks has shown the effectiveness of the templates and that BERTSubs can dramatically outperform the baselines that use (literal-aware) knowledge graph embeddings, non-contextual word embeddings and the state-of-the-art OWL ontology embeddings.




Similar content being viewed by others
Data Availability
All the data used in the experiments have been open. The materials used in the paper, such as the figures, will also be open. The codes used in the experiments have been open. They will be further maintained and kept open as a part of the DeepOnto library.
Notes
obo: is short for the prefix of http://www.geneontology.org/formats/oboInOwl#.
Our codes and data: https://gitlab.com/chen00217/bert_subsumption
ncit: denotes the prefix of http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#
References
Baader, F., Brandt, S., Lutz, C.: Pushing the EL envelope. IJCAI 5, 364–369 (2005)
Baader, F., Horrocks, I., Lutz, C., Sattler, U.: Introduction to description logic. Cambridge University Press (2017)
Bechhofer, S., Van Harmelen, F., Hendler, J., Horrocks, I., McGuinness, D.L., Patel-Schneider, P.F., Stein, L.A., et al.: OWL web ontology language reference. W3C Recommendation 10(2), 1–53 (2004)
Bordes, A., Usunier, N., Garcia-Duran, A., Weston, J., Yakhnenko, O.: Translating embeddings for modeling multi-relational data. Advances in Neural Information Processing Systems 26 (2013)
Bosselut, A., Rashkin, H., Sap, M., Malaviya, C., Celikyilmaz, A., Choi, Y.: COMET: Commonsense transformers for automatic knowledge graph construction. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. pp. 4762–4779 (2019)
Chen, J., Hu, P., Jimenez-Ruiz, E., Holter, O.M., Antonyrajah, D., Horrocks, I.: OWL2Vec*: Embedding of OWL ontologies. Machine Learning pp. 1–33 (2021)
Chen, J., Jiménez-Ruiz, E., Horrocks, I., Antonyrajah, D., Hadian, A., Lee, J.: Augmenting ontology alignment by semantic embedding and distant supervision. In: European Semantic Web Conference. pp. 392–408. Springer (2021)
Consortium, G.O.: The gene ontology project in 2008. Nucleic acids research 36(suppl_1), D440–D444 (2008)
Cuenca Grau, B., Horrocks, I., Motik, B., Parsia, B., Patel-Schneider, P.F., Sattler, U.: OWL 2: The next step for OWL. J. Web Semantics 6(4), 309–322 (2008)
Devlin, J., Chang, M.W., Lee, K., Toutanova, K.: BERT: Pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). pp. 4171–4186 (2019)
Dooley, D.M., Griffiths, E.J., Gosal, G.S., Buttigieg, P.L., Hoehndorf, R., Lange, M.C., Schriml, L.M., Brinkman, F.S., Hsiao, W.W.: Foodon: a harmonized food ontology to increase global food traceability, quality control and data integration. npj Sci. Food 2(1), 1–10 (2018)
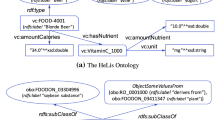
Dragoni, M., Bailoni, T., Maimone, R., Eccher, C.: HeLiS: An ontology for supporting healthy lifestyles. In: International semantic web conference. pp. 53–69. Springer (2018)
Ebrahimi, M., Eberhart, A., Bianchi, F., Hitzler, P.: Towards bridging the neuro-symbolic gap: Deep deductive reasoners. Appl. Intell. 51(9), 6326–6348 (2021)
Gao, T., Fisch, A., Chen, D.: Making pre-trained language models better few-shot learners. In: Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing. pp. 3816–3830 (2021)
Garg, D., Ikbal, S., Srivastava, S.K., Vishwakarma, H., Karanam, H., Subramaniam, L.V.: Quantum embedding of knowledge for reasoning. Advances in Neural Information Processing Systems 32 (2019)
Gesese, G.A., Biswas, R., Alam, M., Sack, H.: A survey on knowledge graph embeddings with literals: Which model links better literal-ly? Semantic Web pp. 1–31 (2019)
Glimm, B., Horrocks, I., Motik, B., Stoilos, G., Wang, Z.: HermiT: an OWL 2 reasoner. J. Autom. Reason. 53(3), 245–269 (2014)
He, Y., Chen, J., Antonyrajah, D., Horrocks, I.: BERTMap: A BERT-based ontology alignment system. In: AAAI (2022)
He, Y., Chen, J., Dong, H., Jiménez-Ruiz, E., Hadian, A., Horrocks, I.: Machine learning-friendly biomedical datasets for equivalence and subsumption ontology matching. In: The Semantic Web–ISWC 2022: 21st International Semantic Web Conference, Virtual Event, October 23–27, 2022, Proceedings. pp. 575–591. Springer (2022)
Horrocks, I., Chen, J., Lee, J.: Tool support for ontology design and quality assurance. In: ICBO 2020 integrated food ontology workshop (IFOW) (2020)
Horrocks, I., Patel-Schneider, P.F., Boley, H., Tabet, S., Grosof, B., Dean, M., et al.: SWRL: A semantic web rule language combining OWL and RuleML. W3C Member submission 21(79), 1–31 (2004)
Kaljurand, K.: Attempto controlled english as a semantic web language. University of Tartu (2007)
Kazakov, Y., Krötzsch, M., Simančík, F.: The incredible ELK. J. Autom. Reason 53(1), 1–61 (2014)
Kulmanov, M., Liu-Wei, W., Yan, Y., Hoehndorf, R.: EL embeddings: Geometric construction of models for the description logic EL++. In: IJCAI (2019)
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C.H., Kang, J.: BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36(4), 1234–1240 (2020)
Lees, A., Welty, C., Zhao, S., Korycki, J., Mc Carthy, S.: Embedding semantic taxonomies. In: Proceedings of the 28th International Conference on Computational Linguistics. pp. 1279–1291 (2020)
Lin, Y., Liu, Z., Sun, M., Liu, Y., Zhu, X.: Learning entity and relation embeddings for knowledge graph completion. In: Twenty-ninth AAAI conference on artificial intelligence (2015)
Liu, H., Perl, Y., Geller, J.: Concept placement using bert trained by transforming and summarizing biomedical ontology structure. J. Biomed. Inform. 112, 103607 (2020)
Liu, P., Yuan, W., Fu, J., Jiang, Z., Hayashi, H., Neubig, G.: Pre-train, prompt, and predict: A systematic survey of prompting methods in natural language processing. arXiv:2107.13586 (2021)
Mikolov, T., Chen, K., Corrado, G., Dean, J.: Efficient estimation of word representations in vector space. arXiv:1301.3781 (2013)
Mousselly-Sergieh, H., Botschen, T., Gurevych, I., Roth, S.: A multimodal translation-based approach for knowledge graph representation learning. In: Proceedings of the Seventh Joint Conference on Lexical and Computational Semantics. pp. 225–234 (2018)
Musen, M.A.: The protégé project: a look back and a look forward. AI Matters 1(4), 4–12 (2015)
Nickel, M., Kiela, D.: Poincaré embeddings for learning hierarchical representations. Adv. Neural Inform. Process. Syst. 30, 6338–6347 (2017)
Ochs, C., Geller, J., Perl, Y., Musen, M.A.: A unified software framework for deriving, visualizing, and exploring abstraction networks for ontologies. J. Biomed. Inform. 62, 90–105 (2016)
Petroni, F., Rocktäschel, T., Riedel, S., Lewis, P., Bakhtin, A., Wu, Y., Miller, A.: Language models as knowledge bases? In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing. pp. 2463–2473 (2019)
Raffel, C., Shazeer, N., Roberts, A., Lee, K., Narang, S., Matena, M., Zhou, Y., Li, W., Liu, P.J.: Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21, 1–67 (2020)
Schriml, L.M., Mitraka, E., Munro, J., Tauber, B., Schor, M., Nickle, L., Felix, V., Jeng, L., Bearer, C., Lichenstein, R., Bisordi, K., Campion, N., Hyman, B., Kurland, D., Oates, C.P., Kibbey, S., Sreekumar, P., Le, C., Giglio, M., Greene, C.: Human Disease Ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Research (2018)
Schuster, M., Nakajima, K.: Japanese and Korean voice search. In: 2012 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). pp. 5149–5152. IEEE (2012)
Sioutos, N., de Coronado, S., Haber, M.W., Hartel, F.W., Shaiu, W.L., Wright, L.W.: NCI Thesaurus: A semantic model integrating cancer-related clinical and molecular information. J. Biomed. Inform. 40(1), 30–43 (2007), bio*Medical Informatics
Smaili, F.Z., Gao, X., Hoehndorf, R.: Onto2Vec: Joint vector-based representation of biological entities and their ontology-based annotations. Bioinformatics 34(13), i52–i60 (2018)
Smaili, F.Z., Gao, X., Hoehndorf, R.: OPA2Vec: Combining formal and informal content of biomedical ontologies to improve similarity-based prediction. Bioinformatics 35(12), 2133–2140 (2019)
Soylu, A., Kharlamov, E., Zheleznyakov, D., Jimenez-Ruiz, E., Giese, M., Skjæveland, M.G., Hovland, D., Schlatte, R., Brandt, S., Lie, H., et al.: Optiquevqs: A visual query system over ontologies for industry. Semantic Web 9(5), 627–660 (2018)
Staab, S., Studer, R.: Handbook on ontologies. Springer Science & Business Media (2010)
Stevens, R., Malone, J., Williams, S., Power, R., Third, A.: Automating generation of textual class definitions from OWL to English. In: Journal of Biomedical Semantics. vol. 2, pp. 1–20. Springer (2011)
Vilnis, L., Li, X., Murty, S., McCallum, A.: Probabilistic embedding of knowledge graphs with box lattice measures. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). pp. 263–272 (2018)
Wang, B., Shen, T., Long, G., Zhou, T., Wang, Y., Chang, Y.: Structure-augmented text representation learning for efficient knowledge graph completion. In: Proceedings of the Web Conference 2021. pp. 1737–1748 (2021)
Wang, Q., Mao, Z., Wang, B., Guo, L.: Knowledge graph embedding: A survey of approaches and applications. IEEE Trans. Knowl. Data Eng. 29(12), 2724–2743 (2017)
Wu, Y., Schuster, M., Chen, Z., Le, Q.V., Norouzi, M., Macherey, W., Krikun, M., Cao, Y., Gao, Q., Macherey, K., et al.: Google’s neural machine translation system: Bridging the gap between human and machine translation. arXiv:1609.08144 (2016)
Xiong, B., Potyka, N., Tran, T.K., Nayyeri, M., Staab, S.: Box embeddings for the Description Logic EL++. arXiv:2201.09919 (2022)
Yang, B., Yih, W.t., He, X., Gao, J., Deng, L.: Embedding entities and relations for learning and inference in knowledge bases. arXiv:1412.6575 (2014)
Yao, L., Mao, C., Luo, Y.: KG-BERT: BERT for knowledge graph completion. arXiv:1909.03193 (2019)
Zhang, Z., Cai, J., Zhang, Y., Wang, J.: Learning hierarchy-aware knowledge graph embeddings for link prediction. In: Proceedings of the AAAI Conference on Artificial Intelligence. vol. 34, pp. 3065–3072 (2020)
Zhou, L., Cheatham, M., Krisnadhi, A., Hitzler, P.: Geolink data set: A complex alignment benchmark from real-world ontology. Data Intell. 2(3), 353–378 (2020)
Funding
This research was funded in whole or in part by the SIRIUS Centre for Scalable Data Access (Research Council of Norway, project 237889), eBay, Samsung Research UK, Siemens AG, and the EPSRC projects OASIS (EP/S032347/1), UK FIRES (EP/S019111/1) and ConCur (EP/V050869/1). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript (AAM) version arising from this submission
Author information
Authors and Affiliations
Contributions
Jiaoyan Chen is the corresponding author, with major contribution to the paper discussion, experiments and writing. Yuan He and Yuxia Geng contributed partially to the paper discussion and experiments. Ernesto Jimenez-Ruiz, Hang Dong and Ian Horrocks contributed partially to the paper discussion and writing.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable. The paper does not involve any ethics issues.
Conflicts of interest
This paper has conflicts of interest with members from The University of Manchester (manchester.ac.uk), University of Oxford (ox.ac.uk), Zhejiang University (zju.edu.cn), City University of London (city.ac.uk) and University of Oslo (uio.no).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the Topical Collection: Special Issue on Knowledge-Graph-Enabled Methods and Applications for the Future Web Guest Editors: Xin Wang, Jeff Pan, Qingpeng Zhang, and Yuan-Fang Li.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J., He, Y., Geng, Y. et al. Contextual semantic embeddings for ontology subsumption prediction. World Wide Web 26, 2569–2591 (2023). https://doi.org/10.1007/s11280-023-01169-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11280-023-01169-9