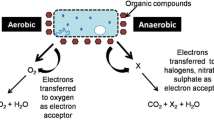
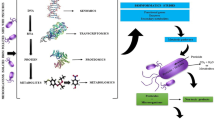
Abstract
Pesticide pollution in recent times has emerged as a grave environmental problem contaminating both aquatic and terrestrial ecosystems owing to their widespread use. Bioremediation using gene editing and system biology could be developed as an eco-friendly and proficient tool to remediate pesticide-contaminated sites due to its advantages and greater public acceptance over the physical and chemical methods. However, it is indispensable to understand the different aspects associated with microbial metabolism and their physiology for efficient pesticide remediation. Therefore, this review paper analyses the different gene editing tools and multi-omics methods in microbes to produce relevant evidence regarding genes, proteins and metabolites associated with pesticide remediation and the approaches to contend against pesticide-induced stress. We systematically discussed and analyzed the recent reports (2015–2022) on multi-omics methods for pesticide degradation to elucidate the mechanisms and the recent advances associated with the behaviour of microbes under diverse environmental conditions. This study envisages that CRISPR-Cas, ZFN and TALEN as gene editing tools utilizing Pseudomonas, Escherichia coli and Achromobacter sp. can be employed for remediation of chlorpyrifos, parathion-methyl, carbaryl, triphenyltin and triazophos by creating gRNA for expressing specific genes for the bioremediation. Similarly, systems biology accompanying multi-omics tactics revealed that microbial strains from Paenibacillus, Pseudomonas putida, Burkholderia cenocepacia, Rhodococcus sp. and Pencillium oxalicum are capable of degrading deltamethrin, p-nitrophenol, chlorimuron-ethyl and nicosulfuron. This review lends notable insights into the research gaps and provides potential solutions for pesticide remediation by using different microbe-assisted technologies. The inferences drawn from the current study will help researchers, ecologists, and decision-makers gain comprehensive knowledge of value and application of systems biology and gene editing in bioremediation assessments.
Graphical Abstract






Similar content being viewed by others
Data availability
Data sharing is not applicable to this article as no new data were created or analysed in this study.
References
Ahmed S, Mofijur M, Nuzhat S, Chowdhury AT, Rafa N, Uddin MA, Inayat A, Mahlia T, Ong HC, Chia WY (2021) Recent developments in physical, biological, chemical, and hybrid treatment techniques for removing emerging contaminants from wastewater. J Hazard Mater 416:125912
Aldas-Vargas A, Poursat BA, Sutton NB (2022) Potential and limitations for monitoring of pesticide biodegradation at trace concentrations in water and soil. World J Microbiol Biotechnol 38(12):1–18
Aliko V, Multisanti CR, Turani B, Faggio C (2022) Get rid of marine pollution: bioremediation an innovative, attractive, and successful cleaning strategy. Sustainability 14(18):11784
Allmaras RR, Wilkins D, Burnside O, Mulla D (2018) Agricultural technology and adoption of conservation practices. Advances in soil and water conservation. Routledge, pp 99–158
Altenhoff AM, Glover NM, Train C-M, Kaleb K, Warwick Vesztrocy A, Dylus D, De Farias TM, Zile K, Stevenson C, Long J (2018) The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res 46(D1):D477–D485
Anjum R, Rahman M, Masood F, Malik A (2012) Bioremediation of pesticides from soil and wastewater. Environmental protection strategies for sustainable development: 295–328
Arazoe T, Kondo A, Nishida K (2018) Targeted nucleotide editing technologies for microbial metabolic engineering. Biotechnol J 13(9):1700596
Arora PK, Bae H (2014) Integration of bioinformatics to biodegradation. Biol procedures online 16(1):1–10
Arraiza Bermudez-Cañete MP, Santamarta Cerezal JC, Ioras F, García Rodríguez JL, Abrudan IV, Korjus H, Borbála G (2014) Climate Change and Restoration of Degraded Land. Colegio de Ingenieros de Montes
Azam S, Parthasarathy S, Singh C, Kumar S, Siddavattam D (2019) Genome organization and adaptive potential of archetypal organophosphate degrading Sphingobium fuliginis ATCC 27551. Genome Biol Evol 11(9):2557–2562
Bala S, Garg D, Thirumalesh BV, Sharma M, Sridhar K, Inbaraj BS, Tripathi M (2022) Recent strategies for bioremediation of emerging pollutants: a review for a green and sustainable environment. Toxics 10(8):484
Banerjee A, Banerjee C, Negi S, Chang J-S, Shukla P (2018) Improvements in algal lipid production: a systems biology and gene editing approach. Crit Rev Biotechnol 38(3):369–385
Bardot C, Besse-Hoggan P, Carles L, Le Gall M, Clary G, Chafey P, Federici C, Broussard C, Batisson I (2015) How the edaphic Bacillus megaterium strain Mes11 adapts its metabolism to the herbicide mesotrione pressure. Environ Pollut 199:198–208
Barszczewska-Pietraszek G, Drzewiecka M, Czarny P, Skorski T, Śliwiński T (2022) Polθ Inhibition: an anticancer therapy for HR-Deficient tumours. Int J Mol Sci 24(1):319
Basu S, Rabara RC, Negi S, Shukla P (2018) Engineering PGPMOs through gene editing and systems biology: a solution for phytoremediation? Trends Biotechnol 36(5):499–510
Bernat P, Nykiel-Szymańska J, Stolarek P, Słaba M, Szewczyk R, Różalska S (2018) 2, 4-dichlorophenoxyacetic acid-induced oxidative stress: Metabolome and membrane modifications in Umbelopsis isabellina, a herbicide degrader. PLoS ONE 13(6):e0199677
Bhardwaj A, Nain V (2021) TALENs—an indispensable tool in the era of CRISPR: a mini review. J Genetic Eng Biotechnol 19(1):1–10
Bhatt P, Pathak VM, Joshi S, Bisht TS, Singh K, Chandra D (2019) Major metabolites after degradation of xenobiotics and enzymes involved in these pathways. Smart bioremediation technologies. Elsevier, pp 205–215
Bhatt P, Bhandari G, Bhatt K, Maithani D, Mishra S, Gangola S, Bhatt R, Huang Y, Chen S (2021a) Plasmid-mediated catabolism for the removal of xenobiotics from the environment. J Hazard Mater 420:126618
Bhatt P, Bhatt K, Sharma A, Zhang W, Mishra S, Chen S (2021b) Biotechnological basis of microbial consortia for the removal of pesticides from the environment. Crit Rev Biotechnol 41(3):317–338
Bhatt P, Bhatt K, Huang Y, Li J, Wu S, Chen S (2022) Biofilm formation in xenobiotic-degrading microorganisms.Critical Reviews in Biotechnology:1–21
Bhatt P, Bhatt K, Chen W-J, Huang Y, Xiao Y, Wu S, Lei Q, Zhong J, Zhu X, Chen S (2023) Bioremediation potential of laccase for catalysis of glyphosate, isoproturon, lignin, and parathion: molecular docking, dynamics, and simulation. J Hazard Mater 443:130319
Bier E, Harrison MM, O’Connor-Giles KM, Wildonger J (2018) Advances in engineering the fly genome with the CRISPR-Cas system. Genetics 208(1):1–18
Bourguignon N, Irazusta V, Isaac P, Estévez C, Maizel D, Ferrero MA (2019) Identification of proteins induced by polycyclic aromatic hydrocarbon and proposal of the phenanthrene catabolic pathway in Amycolatopsis tucumanensis DSM 45259. Ecotoxicol Environ Saf 175:19–28
Broeders M, Herrero-Hernandez P, Ernst MP, van der Ploeg AT, Pijnappel WP (2020) Sharpening the molecular scissors: advances in gene-editing technology. Iscience 23(1):100789
Cabral L, Giovanella P, Pellizzer EP, Teramoto EH, Kiang CH, Sette LD (2022) Microbial communities in petroleum-contaminated sites: structure and metabolisms. Chemosphere 286:131752
Campe R, Hollenbach E, Kämmerer L, Hendriks J, Höffken HW, Kraus H, Lerchl J, Mietzner T, Tresch S, Witschel M (2018) A new herbicidal site of action: cinmethylin binds to acyl-ACP thioesterase and inhibits plant fatty acid biosynthesis. Pestic Biochem Physiol 148:116–125
Canver MC, Joung JK, Pinello L (2018) Impact of genetic variation on CRISPR-Cas targeting. CRISPR J 1(2):159–170
Castrejón-Godínez ML, Ortiz-Hernández ML, Salazar E, Encarnación S, Mussali-Galante P, Tovar-Sánchez E, Sánchez-Salinas E, Rodríguez A (2019) Transcriptional analysis reveals the metabolic state of Burkholderia zhejiangensis CEIB S4-3 during methyl parathion degradation. PeerJ 7:e6822
Castrejón-Godínez ML, Tovar-Sánchez E, Ortiz-Hernández ML, Encarnación-Guevara S, Martínez-Batallar ÁG, Hernández-Ortiz M, Sánchez-Salinas E, Rodríguez A, Mussali-Galante P (2022) Proteomic analysis of Burkholderia zhejiangensis CEIB S4–3 during the methyl parathion degradation process. Pestic Biochem Physiol 187:105197
Chakraborty R, Wu CH, Hazen TC (2012) Systems biology approach to bioremediation. Curr Opin Biotechnol 23(3):483–490
Chandran H, Meena M, Sharma K (2020) Microbial biodiversity and bioremediation assessment through omics approaches. Front Environ Chem 1:570326
Chatterjee G, Negi S, Basu S, Faintuch J, O’Donovan A, Shukla P (2022) Microbiome systems biology advancements for natural well-being. Science of The Total Environment:155915
Chen S, Deng Y, Chang C, Lee J, Cheng Y, Cui Z, Zhou J, He F, Hu M, Zhang L-H (2015) Pathway and kinetics of cyhalothrin biodegradation by Bacillus thuringiensis strain ZS-19. Sci Rep 5(1):1–10
Chen Q, Tu H, Luo X, Zhang B, Huang F, Li Z, Wang J, Shen W, Wu J, Cui Z (2016) The regulation of para-nitrophenol degradation in Pseudomonas putida DLL-E4. PLoS ONE 11(5):e0155485
Chen ZJ, Zhai XY, Liu J, Zhang N, Yang H (2022) Detoxification and catabolism of mesotrione and fomesafen facilitated by a phase II reaction acetyltransferase in rice. Journal of Advanced Research
Cheng Y, Zang H, Wang H, Li D, Li C (2018) Global transcriptomic analysis of Rhodococcus erythropolis D310-1 in responding to chlorimuron-ethyl. Ecotoxicol Environ Saf 157:111–120
Chimekagbe OE, Amadi CF, Okolonkwo BN (2022) Organophosphates toxicity: pathophysiology, diagnosis, and treatment.Asian Journal of Biochemistry, Genetics and Molecular Biology:27–37
Cooper LA, Stringer AM, Wade JT (2018) Determining the specificity of cascade binding, interference, and primed adaptation in vivo in the Escherichia coli type IE CRISPR-Cas system. MBio 9(2):e02100–02117
Cuperlovic-Culf M (2018) Machine learning methods for analysis of metabolic data and metabolic pathway modeling. Metabolites 8(1):4
Dai Z, Zhang S, Yang Q, Zhang W, Qian X, Dong W, Jiang M, Xin F (2018) Genetic tool development and systemic regulation in biosynthetic technology. Biotechnol Biofuels 11(1):1–12
Dangi AK, Sharma B, Hill RT, Shukla P (2019) Bioremediation through microbes: systems biology and metabolic engineering approach. Crit Rev Biotechnol 39(1):79–98
Dash DM, Osborne WJ (2022) A systematic review on the implementation of advanced and evolutionary biotechnological tools for efficient bioremediation of organophosphorus pesticides. Chemosphere:137506
De Sousa CS, Hassan SS, Pinto AC, Silva WM, De Almeida SS, Soares SDC, Azevedo MS, Rocha CS, Barh D, Azevedo V (2018) Microbial omics: applications in biotechnology. Omics technologies and bio-engineering. Elsevier, pp 3–20
Dhananjayan V, Jayanthi P, Ravichandran B, Jayakumar R (2022) Biomonitoring and biomarkers of pesticide exposure and human health risk assessment.Pesticides in the Natural Environment:563–584
Drangert J-O, Tonderski K, McConville J (2018) Extending the european Union Waste Hierarchy to Guide nutrient-effective urban sanitation toward global food security—Opportunities for phosphorus recovery. Front Sustainable Food Syst 2:3
Dreher J, Scheiber J, Stiefl N, Baumann K (2018) xMaP An interpretable alignment-free four-dimensional quantitative structure–activity relationship technique based on Molecular Surface Properties and Conformer Ensembles. J Chem Inf Model 58(1):165–181
Duarte M, Nielsen A, Camarinha-Silva A, Vilchez‐Vargas R, Bruls T, Wos‐Oxley ML, Jauregui R, Pieper DH (2017) Functional soil metagenomics: elucidation of polycyclic aromatic hydrocarbon degradation potential following 12 years of in situ bioremediation. Environ Microbiol 19(8):2992–3011
El Amrani A, Dumas A-S, Wick LY, Yergeau E, Berthomé R (2015) Omics” insights into PAH degradation toward improved green remediation biotechnologies. Environ Sci Technol 49(19):11281–11291
Elmassry MM, Zayed A, Farag MA (2022) Gut homeostasis and microbiota under attack: impact of the different types of food contaminants on gut health. Crit Rev Food Sci Nutr 62(3):738–763
Erythropel HC, Zimmerman JB, de Winter TM, Petitjean L, Melnikov F, Lam CH, Lounsbury AW, Mellor KE, Janković NZ, Tu Q (2018) The Green ChemisTREE: 20 years after taking root with the 12 principles. Green Chem 20(9):1929–1961
Eykelbosh A (2014) Short-and long-term health impacts of marine and terrestrial oil spills. Vancouver Coastal Health
Fang L-C, Chen Y-F, Wang D-S, Sun L-N, Tang X-Y, Hua R-M (2016) Complete genome sequence of a novel chlorpyrifos degrading bacterium, Cupriavidus nantongensis X1. J Biotechnol 227:1–2
Feng W, Wei Z, Song J, Qin Q, Yu K, Li G, Zhang J, Wu W, Yan Y (2017) Hydrolysis of nicosulfuron under acidic environment caused by oxalate secretion of a novel Penicillium oxalicum strain YC-WM1. Sci Rep 7(1):1–11
Ferreira P, Choupina AB (2022) CRISPR/Cas9 a simple, inexpensive and effective technique for gene editing. Mol Biol Rep 49(7):7079–7086
Fu R, Wang Y, Liu Y, Liu H, Zhao Q, Zhang Y, Wang C, Li Z, Jiao B, He Y (2022) CRISPR-Cas12a based fluorescence assay for organophosphorus pesticides in agricultural products. Food Chem 387:132919
Gangola S, Khati P, Sharma A (2015) Mycoremediation of imidaclopridin the presence of different soil amendments using Trichoderma longibrachiatum and aspergillus oryzae isolated from pesticide contaminated agricultural fields of Uttarakhand. J Bioremediat Biodegr 6(5):1
Gangola S, Sharma A, Bhatt P, Khati P, Chaudhary P (2018) Presence of esterase and laccase in Bacillus subtilis facilitates biodegradation and detoxification of cypermethrin. Sci Rep 8(1):12755
Gangola S, Joshi S, Kumar S, Sharma A (2021a) Differential analysis of pesticides biodegradation in soil using conventional and high-throughput technology. bioRxiv
Gangola S, Joshi S, Kumar S, Sharma B, Sharma A (2021b) Differential proteomic analysis under pesticides stress and normal conditions in Bacillus cereus 2D. PLoS ONE 16(8):e0253106
Gangola S, Bhatt P, Joshi S, Bhandari NS, Kumar S, Prakash O, Kumar A, Tewari S, Nainwal D (2022a) Isolation, Enrichment, and characterization of Fungi for the degradation of Organic Contaminants. In: Mycoremediation Protocols. Springer, pp 1–11
Gangola S, Bhatt P, Kumar AJ, Bhandari G, Joshi S, Punetha A, Bhatt K, Rene ER (2022b) Biotechnological tools to elucidate the mechanism of pesticide degradation in the environment. Chemosphere:133916
Gangola S, Joshi S, Joshi D, Rajwar J, Kour S, Singh J, Kumar S (2022c) Advanced Molecular Technologies for Environmental Restoration and sustainability.Bioremediation of Environmental Pollutants: Emerging Trends and Strategies:385–396
Gangola S, Joshi S, Bhandari G, Bhatt P, Kumar S, Pandey SC (2023) Omics approaches to pesticide biodegradation for sustainable environment. Advanced Microbial Techniques in Agriculture, Environment, and Health Management. Elsevier, pp 191–203
Gaur N, Narasimhulu K, PydiSetty Y (2018) Recent advances in the bio-remediation of persistent organic pollutants and its effect on environment. J Clean Prod 198:1602–1631
Gil FN, Gonçalves AC, Becker JD, Viegas CA (2018) Comparative analysis of transcriptomic responses to sub-lethal levels of six environmentally relevant pesticides in Saccharomyces cerevisiae. Ecotoxicology 27(7):871–889
Giri BS, Geed S, Vikrant K, Lee SS, Kim K-H, Kailasa SK, Vithanage M, Chaturvedi P, Rai BN, Singh RS (2021) Progress in bioremediation of pesticide residues in the environment.Environmental Engineering Research26 (6)
Gong J, Liu X, Cao X, Diao Y, Gao D, Li H, Qian X (2013) PTID: an integrated web resource and computational tool for agrochemical discovery. Bioinformatics 29(2):292–294
Goodwin L, Carra I, Campo P, Soares A (2017) Treatment options for reclaiming wastewater produced by the pesticide industry.Int J Water Wastewater Treat4 (1)
Gosak M, Markovič R, Dolenšek J, Rupnik MS, Marhl M, Stožer A, Perc M (2018) Network science of biological systems at different scales: a review. Phys Life Rev 24:118–135
Gosavi G, Ren B, Li X, Zhou X, Spetz C, Zhou H (2022) A new era in herbicide-tolerant crops development by targeted genome editing. ACS Agricultural Science & Technology 2(2):184–191
Greene AC (2018) CRISPR-based antibacterials: transforming bacterial defense into offense. Trends Biotechnol 36(2):127–130
Guerra AB, Oliveira JS, Silva-Portela RC, Araújo W, Carlos AC, Vasconcelos ATR, Freitas AT, Domingos YS, de Farias MF, Fernandes GJT (2018) Metagenome enrichment approach used for selection of oil-degrading bacteria consortia for drill cutting residue bioremediation. Environ Pollut 235:869–880
Gupta SK, Shukla P (2017) Gene editing for cell engineering: trends and applications. Crit Rev Biotechnol 37(5):672–684
Hao X, Zhang X, Duan B, Huo S, Lin W, Xia X, Liu K (2018) Screening and genome sequencing of deltamethrin-degrading bacterium ZJ6. Curr Microbiol 75(11):1468–1476
Hassan S, Khurshid Z, Bhat SA, Kumar V, Ameen F, Ganai BA (2022) Marine bacteria and omic approaches: a novel and potential repository for bioremediation assessment. J Appl Microbiol 133(4):2299–2313
Ibrahim M, Raajaraam L, Raman K (2021) Modelling microbial communities: harnessing consortia for biotechnological applications. Comput Struct Biotechnol J 19:3892–3907
Imam A, Suman SK, Kanaujia PK, Ray A (2022) Biological machinery for polycyclic aromatic hydrocarbons degradation: a review. Bioresour Technol 343:126121
Jaiswal S, Shukla P (2020) Alternative strategies for microbial remediation of pollutants via synthetic biology. Front Microbiol 11:808
Jaiswal S, Singh DK, Shukla P (2019) Gene editing and systems biology tools for pesticide bioremediation: a review. Front Microbiol 10:87
Jeffries TC, Rayu S, Nielsen UN, Lai K, Ijaz A, Nazaries L, Singh BK (2018) Metagenomic functional potential predicts degradation rates of a model organophosphorus xenobiotic in pesticide contaminated soils. Front Microbiol 9:147
Jokar S, Shavandi M, Haddadi A, Alaie E (2022) Remediation of chlorpyrifos and deltamethrin in different salinity soils and its impact on soil bacterial composition. Int J Environ Sci Technol 19(12):12057–12068
Karahalil B (2016) Overview of systems biology and omics technologies. Curr Med Chem 23(37):4221–4230
Khan Z, Ali Z, Khan AA, Sattar T, Zeshan A, Saboor T, Binyamin B (2022) History and classification of CRISPR/Cas System. The CRISPR/Cas Tool Kit for Genome Editing. Springer, pp 29–52
Kimes NE, Callaghan AV, Aktas DF, Smith WL, Sunner J, Golding B, Drozdowska M, Hazen TC, Suflita JM, Morris PJ (2013) Metagenomic analysis and metabolite profiling of deep–sea sediments from the Gulf of Mexico following the Deepwater Horizon oil spill. Front Microbiol 4:50
Kumari P, Kumar Y (2021) Bioinformatics and computational tools in bioremediation and biodegradation of environmental pollutants. Bioremediation for environmental sustainability. Elsevier, pp 421–444
Laad M, Ghule B (2022) Removal of toxic contaminants from drinking water using biosensors: a systematic review.Groundwater for Sustainable Development:100888
Li M, Song J, Ma Q, Kong D, Zhou Y, Jiang X, Parales R, Ruan Z, Zhang Q (2020) Insight into the characteristics and new mechanism of nicosulfuron biodegradation by a Pseudomonas sp. LAM1902. J Agric Food Chem 68(3):826–837
Li J, Jia C, Lu Q, Hungate BA, Dijkstra P, Wang S, Wu C, Chen S, Li D, Shim H (2021) Mechanistic insights into the success of xenobiotic degraders resolved from metagenomes of microbial enrichment cultures. J Hazard Mater 418:126384
Li J, Chen W-J, Zhang W, Zhang Y, Lei Q, Wu S, Huang Y, Mishra S, Bhatt P, Chen S (2022a) Effects of free or immobilized bacterium Stenotrophomonas acidaminiphila Y4B on glyphosate degradation performance and indigenous microbial community structure. J Agric Food Chem 70(43):13945–13958
Li M, Li Q, Yao J, Sunahara G, Duran R, Zhang Q, Ruan Z (2022b) Transcriptomic response of Pseudomonas nicosulfuronedens LAM1902 to the sulfonylurea herbicide nicosulfuron. Sci Rep 12(1):1–13
Li K, Jia W, Xu L, Zhang M, Huang Y (2023) The plastisphere of biodegradable and conventional microplastics from residues exhibit distinct microbial structure, network and function in plastic-mulching farmland. J Hazard Mater 442:130011
Liu X-Y, Luo X-J, Li C-X, Lai Q-L, Xu J-H (2014) Draft genome sequence of Burkholderia sp. Strain MP-1, a Methyl Parathion (MP)-degrading bacterium from MP-contaminated soil. Genome announcements 2(3):e00344–e00314
Liu J, Wang S, Qin T, Li N, Niu Y, Li D, Yuan Y, Geng H, Xiong L, Liu D (2015) Whole transcriptome analysis of Penicillium digitatum strains treatmented with prochloraz reveals their drug-resistant mechanisms. BMC Genomics 16(1):1–13
Liu Z, Liu Y, Zeng G, Shao B, Chen M, Li Z, Jiang Y, Liu Y, Zhang Y, Zhong H (2018) Application of molecular docking for the degradation of organic pollutants in the environmental remediation: a review. Chemosphere 203:139–150
Liu L, Wu Q, Miao X, Fan T, Meng Z, Chen X, Zhu W (2022) Study on toxicity effects of environmental pollutants based on metabolomics: a review. Chemosphere 286:131815
Luo X-W, Zhang D-Y, Zhu T-H, Zhou X-G, Peng J, Zhang S-B, Liu Y (2018) Adaptation mechanism and tolerance of Rhodopseudomonas palustris PSB-S under pyrazosulfuron-ethyl stress. BMC Microbiol 18(1):1–11
Maier T, Schmidt A, Güell M, Kühner S, Gavin AC, Aebersold R, Serrano L (2011) Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol 7(1):511
Majumdar S, Ligon M, Skinner WC, Terns RM, Terns MP (2017) Target DNA recognition and cleavage by a reconstituted type IG CRISPR-Cas immune effector complex. Extremophiles 21(1):95–107
Malik G, Arora R, Chaturvedi R, Paul MS (2022) Implementation of genetic engineering and novel omics approaches to enhance bioremediation: a focused review. Bull Environ Contam Toxicol 108(3):443–450
Malla MA, Dubey A, Yadav S, Kumar A, Hashem A, Abd_Allah EF (2018) Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front Microbiol 9:1132
Malla MA, Dubey A, Kumar A, Yadav S (2022) Metagenomic analysis displays the potential predictive biodegradation pathways of the persistent pesticides in agricultural soil with a long record of pesticide usage.Microbiological Research:127081
Mandeep, Liu H, Shukla P (2021) Synthetic biology and biocomputational approaches for improving microbial endoglucanases toward their innovative applications. ACS omega 6(9):6055–6063
Mandpe A, Bombaywala S, Paliya S, Kumar S (2022) Efficacy of microbes in removal of pesticides from watershed system. Synergistic approaches for bioremediation of environmental pollutants: recent advances and Challenges. Elsevier, pp 27–51
Meng D, Zhang L, Meng J, Tian Q, Zhai L, Hao Z, Guan Z, Cai Y, Liao X (2019) Evaluation of the strain Bacillus amyloliquefaciens YP6 in Phoxim degradation via transcriptomic data and product analysis. Molecules 24(21):3997
Mesnage R, Oestreicher N, Poirier F, Nicolas V, Boursier C, Vélot C (2020) Transcriptome profiling of the fungus aspergillus nidulans exposed to a commercial glyphosate-based herbicide under conditions of apparent herbicide tolerance. Environ Res 182:109116
Mesnage R, Teixeira M, Mandrioli D, Falcioni L, Ducarmon QR, Zwittink RD, Mazzacuva F, Caldwell A, Halket J, Amiel C (2021) Use of shotgun metagenomics and metabolomics to evaluate the impact of glyphosate or Roundup MON 52276 on the gut microbiota and serum metabolome of Sprague-Dawley rats. Environ Health Perspect 129(1):017005
Mishra S, Lin Z, Pang S, Zhang W, Bhatt P, Chen S (2021a) Recent advanced technologies for the characterization of xenobiotic-degrading microorganisms and microbial communities. Front Bioeng Biotechnol 9:632059
Mishra S, Pang S, Zhang W, Lin Z, Bhatt P, Chen S (2021b) Insights into the microbial degradation and biochemical mechanisms of carbamates. Chemosphere 279:130500
Mitra A, Maitra S (2018) Reproductive toxicity of organophosphate pesticides. Ann Clin Toxicol 2018; 1 (1) 1004
Muangchinda C, Rungsihiranrut A, Prombutara P, Soonglerdsongpha S, Pinyakong O (2018) 16S metagenomic analysis reveals adaptability of a mixed-PAH-degrading consortium isolated from crude oil-contaminated seawater to changing environmental conditions. J Hazard Mater 357:119–127
Naeem M, Majeed S, Hoque MZ, Ahmad I (2020) Latest developed strategies to minimize the off-target effects in CRISPR-Cas-mediated genome editing. Cells 9(7):1608
Nagarajan D, Lee D-J, Varjani S, Lam SS, Allakhverdiev SI, Chang J-S (2022) Microalgae-based wastewater treatment–microalgae-bacteria consortia, multi-omics approaches and algal stress response. Science of the Total Environment 157110
Narayanan M, Ma Y (2022) Influences of biochar on bioremediation/phytoremediation potential of metal-contaminated soils.Frontiers in Microbiology13
Niu A, Lin C (2021) Managing soils of environmental significance: a critical review. J Hazard Mater 417:125990
Okino-Delgado CH, Zanutto-Elgui MR, Prado DZd, Pereira MS, Fleuri LF (2019) Enzymatic bioremediation: current status, challenges of obtaining process, and applications. Microbial metabolism of xenobiotic compounds. Springer, pp 79–101
Olusola AO (2007) Molecular strategies of microbia adaptation to xenobiotics in natural environment. Biotechnol Mol Biology Reviews 2(1):1–13
Olutona G, Fakunle I, Adegbola R (2022) Detection of organochlorine pesticides residue and trace metals in vegetables obtained from Iwo market, Iwo, Nigeria. Int J Environ Sci Technol 19(5):4201–4208
Ortiz-Hernández M, Gama-Martínez Y, Fernández-López M, Castrejón-Godínez ML, Encarnación S, Tovar-Sánchez E, Salazar E, Rodríguez A, Mussali-Galante P (2021) Transcriptomic analysis of Burkholderia cenocepacia CEIB S5-2 during methyl parathion degradation. Environ Sci Pollut Res 28(31):42414–42431
Pan X, Lin D, Zheng Y, Zhang Q, Yin Y, Cai L, Fang H, Yu Y (2016) Biodegradation of DDT by Stenotrophomonas sp. DDT-1: characterization and genome functional analysis. Sci Rep 6(1):1–10
Pan X, Xu T, Xu H, Fang H, Yu Y (2017) Characterization and genome functional analysis of the DDT-degrading bacterium Ochrobactrum sp. DDT-2. Sci Total Environ 592:593–599
Pandey A, Tripathi PH, Tripathi AH, Pandey SC, Gangola S (2019) Omics technology to study bioremediation and respective enzymes. Smart Bioremediation Technologies. Elsevier, pp 23–43
Pankaj GN, Gangola S, Khati P, Kumar G, Srivastava A, Sharma A (2016) Differential expression and characterization of cypermethrin-degrading potential proteins in Bacillus thuringiensis strain, SG4.3 Biotech6 (2)
Panneerselvan L, Krishnan K, Subashchandrabose SR, Naidu R, Megharaj M (2018) Draft genome sequence of Microbacterium esteraromaticum MM1, a bacterium that hydrolyzes the organophosphorus pesticide fenamiphos, isolated from Golf Course soil. Microbiol Resource Announcements 7(4):e00862–e00818
Park H, Seo SI, Lim J-H, Song J, Seo J-H, Kim PI (2022) Screening of Carbofuran-Degrading Bacteria Chryseobacterium sp. BSC2-3 and unveiling the change in Metabolome during Carbofuran Degradation. Metabolites 12(3):219
Patil A, Yesankar P, Bhanse P, Maitreya A, Kapley A, Qureshi A (2022) Omics Perspective: Molecular Blueprint for Agrochemical Bioremediation process in the Environment. Agrochemicals in Soil and Environment. Springer, pp 585–608
Perruchon C, Vasileiadis S, Rousidou C, Papadopoulou ES, Tanou G, Samiotaki M, Garagounis C, Molassiotis A, Papadopoulou KK, Karpouzas DG (2017) Metabolic pathway and cell adaptation mechanisms revealed through genomic, proteomic and transcription analysis of a Sphingomonas haloaromaticamans strain degrading ortho-phenylphenol. Sci Rep 7(1):1–14
Pinu FR, Beale DJ, Paten AM, Kouremenos K, Swarup S, Schirra HJ, Wishart D (2019) Systems biology and multi-omics integration: viewpoints from the metabolomics research community. Metabolites 9(4):76
Plattner J, Kazner C, Naidu G, Wintgens T, Vigneswaran S (2018) Pesticide and microbial contaminants of groundwater and their removal methods: a mini review. J Jaffna Sci Association 1(1):12–18
Purohit HJ, Tikariha H, Kalia VC (2018) Current scenario on application of computational tools in biological systems. In: Soft computing for biological systems. Springer, pp 1–12
Qiao Y, Zhang N, Liu J, Yang H (2023) Interpretation of ametryn biodegradation in rice based on joint analyses of transcriptome, metabolome and chemo-characterization. J Hazard Mater 445:130526
Rafeeq H, Afsheen N, Rafique S, Arshad A, Intisar M, Hussain A, Bilal M, Iqbal HM (2023) Genetically engineered microorganisms for environmental remediation. Chemosphere 310:136751
Rajesh T, Rajendhran J, Gunasekaran P (2012) Genomic technologies in environmental bioremediation. Microorganisms in environmental management:701–718
Razzaq A, Saleem F, Kanwal M, Mustafa G, Yousaf S, Imran Arshad HM, Hameed MK, Khan MS, Joyia FA (2019) Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int J Mol Sci 20(16):4045
Rico E, Jeacock L, Kovářová J, Horn D (2018) Inducible high-efficiency CRISPR-Cas9-targeted gene editing and precision base editing in African trypanosomes. Sci Rep 8(1):1–10
Ridl J, Suman J, Fraraccio S, Hradilova M, Strejcek M, Cajthaml T, Zubrova A, Macek T, Strnad H, Uhlik O (2018) Complete genome sequence of Pseudomonas alcaliphila JAB1 (= DSM 26533), a versatile degrader of organic pollutants. Stand genomic Sci 13(1):1–10
Rodríguez A, Castrejón-Godínez ML, Salazar-Bustamante E, Gama-Martínez Y, Sánchez-Salinas E, Mussali-Galante P, Tovar-Sánchez E, Ortiz-Hernández ML (2020) Omics approaches to pesticide biodegradation. Curr Microbiol 77:545–563
Rodríguez A, Castrejón-Godínez ML, Sánchez-Salinas E, Mussali-Galante P, Tovar-Sánchez E, Ortiz-Hernández M (2022) Pesticide Bioremediation: OMICs Technologies for understanding the processes. In: Pesticides Bioremediation. Springer, pp 197–242
Roy D, Tomo S, Purohit P, Setia P (2021) Microbiome in death and beyond: current vistas and future trends. Front Ecol Evol 9:630397
Sahoo S, Routray SP, Lenka S, Bhuyan R, Mohanty JN (2022) CRISPR/Cas-Mediated functional gene editing for improvement in bioremediation: an emerging strategy. Omics Insights in Environmental Bioremediation. Springer, pp 635–664
Saran LM, Buzzo BB, Sales CR, Alves LMC, Ribeiro RLA (2022) In: Bioremediation (ed) Bioremediation of potentially toxic Metals by Microorganisms and Biomolecules. CRC Press, pp 83–106
Sarker A, Nandi R, Kim J-E, Islam T (2021) Remediation of chemical pesticides from contaminated sites through potential microorganisms and their functional enzymes: prospects and challenges. Environ Technol Innov 23:101777
Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A (2019) The global burden of pathogens and pests on major food crops. Nat Ecol Evol 3(3):430–439
Sengupta K, Alam M, Pailan S, Saha P (2019) Biodegradation of 4-nitrophenol by a Rhodococcus species and a preliminary insight into its toxicoproteome based on mass spectrometry analysis. J Environ Biol 40(3):356–362
Sequino G, Valentino V, Villani F, De Filippis F (2022) Omics-based monitoring of microbial dynamics across the food chain for the improvement of food safety and quality.Food Research International:111242
Shah T, Andleeb T, Lateef S, Noor MA (2018) Genome editing in plants: advancing crop transformation and overview of tools. Plant Physiol Biochem 131:12–21
Shapiro RS, Chavez A, Collins JJ (2018) CRISPR-based genomic tools for the manipulation of genetically intractable microorganisms. Nat Rev Microbiol 16(6):333–339
Shekhar SK, Godheja J, Modi DR (2020) Molecular technologies for assessment of bioremediation and characterization of microbial communities at pollutant-contaminated sites. Bioremediation of industrial waste for environmental safety. Springer, pp 437–474
Sheng Y, Benmati M, Guendouzi S, Benmati H, Yuan Y, Song J, Xia C, Berkani M (2022) Latest eco-friendly approaches for pesticides decontamination using microorganisms and consortia microalgae: a comprehensive insights, challenges, and perspectives. Chemosphere:136183
Shi Y-h, Ren L, Jia Y, Yan Y-c (2015) Genome sequence of organophosphorus pesticide-degrading bacterium Pseudomonas stutzeri strain YC-YH1. Genome Announcements 3(2):e00192–e00115
Singh A, Dhiman N, Kar AK, Singh D, Purohit MP, Ghosh D, Patnaik S (2020) Advances in controlled release pesticide formulations: prospects to safer integrated pest management and sustainable agriculture. J Hazard Mater 385:121525
Singh AK, Bilal M, Iqbal HM, Raj A (2021) Trends in predictive biodegradation for sustainable mitigation of environmental pollutants: recent progress and future outlook. Sci Total Environ 770:144561
Soman R, Kavitha M, Shaji H (2022) Metagenomics: a genomic tool for monitoring microbial communities during bioremediation. Microbes and Microbial Biotechnology for Green Remediation. Elsevier, pp 813–821
Sule RO, Condon L, Gomes AV (2022) A common feature of pesticides: oxidative stress—the role of oxidative stress in pesticide-induced toxicity. Oxidative Medicine and Cellular Longevity 2022
Szewczyk R, Soboń A, Słaba M, Długoński J (2015) Mechanism study of alachlor biodegradation by Paecilomyces marquandii with proteomic and metabolomic methods. J Hazard Mater 291:52–64
Taha EA, Lee J, Hotta A (2022) Delivery of CRISPR-Cas tools for in vivo genome editing therapy: Trends and challenges. Journal of Controlled Release
Tarla DN, Erickson LE, Hettiarachchi GM, Amadi SI, Galkaduwa M, Davis LC, Nurzhanova A, Pidlisnyuk V (2020) Phytoremediation and bioremediation of pesticide-contaminated soil. Appl Sci 10(4):1217
Techtmann SM, Hazen TC (2016) Metagenomic applications in environmental monitoring and bioremediation. J Ind Microbiol Biotechnol 43(10):1345–1354
Thakkar H, Vincent V (2021) Application of Microorganisms in Bioremediation.Bioprospecting of Microorganism-Based Industrial Molecules:77–103
Tiwari B, Verma E, Chakraborty S, Srivastava AK, Mishra AK (2018) Tolerance strategies in cyanobacterium Fischerella sp. under pesticide stress and possible role of a carbohydrate-binding protein in the metabolism of methyl parathion (MP). Int Biodeterior Biodegrad 127:217–226
Tripathi S, Srivastava P, Devi RS, Bhadouria R (2020) Influence of synthetic fertilizers and pesticides on soil health and soil microbiology. Agrochemicals detection, treatment and remediation. Elsevier, pp 25–54
Tudi M, Daniel Ruan H, Wang L, Lyu J, Sadler R, Connell D, Chu C, Phung DT (2021) Agriculture development, pesticide application and its impact on the environment. Int J Environ Res Public Health 18(3):1112
Varga B, Somogyi V, Meiczinger M, Kováts N, Domokos E (2019) Enzymatic treatment and subsequent toxicity of organic micropollutants using oxidoreductases-A review. J Clean Prod 221:306–322
Vasseghian Y, Arunkumar P, Joo S-W, Gnanasekaran L, Kamyab H, Rajendran S, Balakrishnan D, Chelliapan S, Klemeš JJ (2022) Metal-organic framework-enabled pesticides are an emerging tool for sustainable cleaner production and environmental hazard reduction.Journal of Cleaner Production:133966
Vogel C, Marcotte EM (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13(4):227–232
Wang T, Hu C, Zhang R, Sun A, Li D, Shi X (2019a) Mechanism study of cyfluthrin biodegradation by Photobacterium ganghwense with comparative metabolomics. Appl Microbiol Biotechnol 103(1):473–488
Wang W, Chen X, Yan H, Hu J, Liu X (2019b) Complete genome sequence of the cyprodinil-degrading bacterium Acinetobacter johnsonii LXL_C1. Microb Pathog 127:246–249
Wei X, Pu A, Liu Q, Hou Q, Zhang Y, An X, Long Y, Jiang Y, Dong Z, Wu S (2022) The bibliometric landscape of gene editing innovation and regulation in the worldwide. Cells 11(17):2682
Wolfand JM, LeFevre GH, Luthy RG (2016) Metabolization and degradation kinetics of the urban-use pesticide fipronil by white rot fungus Trametes versicolor. Environ Science: Processes Impacts 18(10):1256–1265
Wong DW (2018) Gene targeting and genome editing. The ABCs of Gene Cloning. Springer, pp 187–197
Wu X, Chen W-J, Lin Z, Huang Y, El Sebai TN, Alansary N, El-Hefny DE, Mishra S, Bhatt P, Lü H (2023) Rapid Biodegradation of the Organophosphorus Insecticide Acephate by a novel strain Burkholderia sp. A11 and its impact on the structure of the Indigenous Microbial Community. Journal of Agricultural and Food Chemistry
Yan C, Shi G, Chen J (2022) Fluorescent detection of two pesticides based on CRISPR-Cas12a and its application for the construction of four molecular logic gates. J Agric Food Chem 70(39):12700–12707
Yang H, Hu S, Wang X, Chuang S, Jia W, Jiang J (2020) Pigmentiphaga sp. strain D-2 uses a novel amidase to initiate the catabolism of the neonicotinoid insecticide acetamiprid. Appl Environ Microbiol 86(6):e02425–e02419
Yi W, Yang K, Ye J, Long Y, Ke J, Ou H (2017) Triphenyltin degradation and proteomic response by an engineered Escherichia coli expressing cytochrome P450 enzyme. Ecotoxicol Environ Saf 137:29–34
Zhang T, Tang J, Sun J, Yu C, Liu Z, Chen J (2015) Hex1-related transcriptome of Trichoderma atroviride reveals expression patterns of ABC transporters associated with tolerance to dichlorvos. Biotechnol Lett 37(7):1421–1429
Zhang S, Hu Z, Wang H (2018) A retrospective review of microbiological methods applied in studies following the Deepwater Horizon oil spill. Front Microbiol 9:520
Zhang L, Zhou X-Y, Su X-J, Hu Q, Jiang J-D (2019) Spirosoma sordidisoli sp. nov., a propanil-degrading bacterium isolated from a herbicide-contaminated soil. Antonie Van Leeuwenhoek 112(10):1523–1532
Zhang H, Yuan X, Xiong T, Wang H, Jiang L (2020a) Bioremediation of co-contaminated soil with heavy metals and pesticides: influence factors, mechanisms and evaluation methods. Chem Eng J 398:125657
Zhang Y, Xu Z, Chen Z, Wang G (2020b) Simultaneous degradation of triazophos, methamidophos and carbofuran pesticides in wastewater using an Enterobacter bacterial bioreactor and analysis of toxicity and biosafety. Chemosphere 261:128054
Zhang D, Zhang Z, Unver T, Zhang B (2021) CRISPR/Cas: a powerful tool for gene function study and crop improvement. J Adv Res 29:207–221
Zhang Y, Geng Y, Li S, Shi T, Ma X, Hua R, Fang L (2023) Efficient knocking out of the Organophosphorus Insecticides Degradation Gene opdB in Cupriavidus nantongensis X1T via CRISPR/Cas9 with Red System. Int J Mol Sci 24(6):6003
Zhao X, Ma F, Feng C, Bai S, Yang J, Wang L (2017) Complete genome sequence of Arthrobacter sp. ZXY-2 associated with effective atrazine degradation and salt adaptation. J Biotechnol 248:43–47
Zhou X, Lei D, Tang J, Wu M, Ye H, Zhang Q (2022) Whole genome sequencing and analysis of fenvalerate degrading bacteria citrobacter freundii CD-9. AMB Express 12(1):1–14
Zhu S, Wang H, Jiang W, Yang Z, Zhou Y, He J, Qiu J, Hong Q (2019) Genome analysis of carbaryl-degrading strain Pseudomonas putida XWY-1. Curr Microbiol 76(8):927–929
Acknowledgements
The authors thank the Centre for Research and Development and Department of Environmental Science, University of Kashmir for providing facilities which together could make the present study possible. The author Shahnawaz Hassan acknowledges the financial support from UGC-MANF for Ph.D. research (MANF UGC beneficiary Code: BININ01669533).
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
SH wrote the manuscript under the supervision of BAG. The author BAG facilitated in reviewing and editing the manuscript. SH made the figures for the manuscript using Bio render software.
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that there is no conflict of interest in this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hassan, S., Ganai, B.A. Deciphering the recent trends in pesticide bioremediation using genome editing and multi-omics approaches: a review. World J Microbiol Biotechnol 39, 151 (2023). https://doi.org/10.1007/s11274-023-03603-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03603-6