Abstract
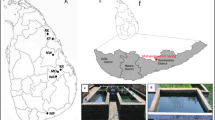
Hot environments are between the supporting life extreme niches that appear to have maintained some degree of pristine quality and of special biotechnological interest. Knowledge on biodiversity in terrestrial hot springs is still scanty and has not been compared in the light of the specificity of those extreme ecological niches. Study on diversity of thermophilic bacteria inhabiting a hot spring located in Rupi Basin (RB), South-West Bulgaria, revealed a high phylogenetic richness in it (genotypic diversity is 0.37). A total of 120 clones were examined, and grouped in 28 phylogenetic types by their RFLP profile. 16S rRNA gene analysis allowed the identification of nine divisions from the domain Bacteria and one Candidate division. Ten of the retrieved bacterial sequences representing one third of the sequence types showed less than 97% similarity to the closest neighbor and were referred as new sequences. Four of them were distantly related to validly described bacteria (showed ≤90% similarity) suggesting new taxons on at least genus level. Comparison of biodiversity in the spring from Rupi Basin, Bulgaria with that described from other terrestrial hot springs revealed that Proteobacteria, Hydrogenobacter/Aquifex and Thermus are common bacterial groups for terrestrial hot springs. Simultaneously, specific bacterial taxons were observed in different springs.


Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Atanassova M, Derekova A, Mandeva R, Sjøholm C, Kambourova M (2008) Anoxybacllus bogrovensis sp. nov., a novel thermophilic bacterium isolated from Dolni Bogrov’s hot spring, Bulgaria. Int J Syst Evol Microbiol 58:2330–2335
Atkinson T, Cairns S, Cowan DA, Danson MJ, Hough DW, Johnson DB, Norris PR, Raven N, Robinson C, Robson R, Sharp RJ (2000) A microbiological survey of Montserrat Island hydrothermal biotopes. Extremophiles 4:305–313
Bojadgieva K, Dipchikova S, Benderev A, Koseva J (2002) Thermal waters and balneology in Bulgaria. GHC Bull 23:18–25
Burggraf S, Olsen GJ, Woese CR (1992) A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol 15:352–356
Derekova A, Sjøholm C, Mandeva R, Kambourova M (2007) Anoxybacillus rupiensis sp. nov., a novel thermophilic bacterium isolated from Rupi basin (Bulgaria). Extremophiles 11:577–583
Ferrera I, Longhorn S, Banta AB, Liu Y, Preston D, Reysenbach AL (2007) Diversity of 16S rRNA gene, ITS region and aclB gene of the Aquificales. Extremophiles 11:57–64
Ferris MJ, Ward DM (1997) Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol 63:1375–1381
Ferris MJ, Nold SC, Santegoeds CM, Ward DM (2001) Examining bacterial population diversity within the Octopus Spring microbial mat community. In: Reysenback A-L, Voytek M, Mancinelli R (eds) Thermophiles: biodiversity, ecology and evolution. Kluwer/Plenum, NY, pp 51–64
Ghosh D, Bal B, Kashyap VK, Pal S (2003) Molecular phylogenetic exploration of bacterial diversity in a Bakreshwar (India) hot spring and culture of Shewanella-related thermophiles. Appl Environ Microbiol 69:4332–4336
Huber R, Huber H, Stetter KO (2000) Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol Rev 24:615–623
Hugenholtz P, Pitulle C, Hershberger KL, Pace NR (1998) Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol 180:366–376
Maidak BL, Larsen N, McCaughey MJ, Overbeek R, Olsen GJ, Fogel K, Blandy J, Woese CR (1994) The ribosomal database project. Nucleic Acids Res 22:3485–3487
Marteinsson VT, Hauksdottir S, Hobel CFV, Kristmannsdottir H, Hreggvidsson GO, Kristiansson JK (2001) Phylogenetic diversity analysis of subterranean hot springs in Iceland. Appl Environ Microbiol 67:4242–4248
Moyer CL, Dobbs FC, Karl DM (1995) Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol 61:1555–1562
Pitulle C, Yang Y, Marchiani M, Moore ERB, Siefert JL, Arano M, Justhuk P, Fox GE (1994) Phylogenetic position of the genus Hydrogenobacter. Int J Syst Bacteriol 44:620–626
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Posada D, Crandall KA (1998) Model test: testing the model of DNA substitution. Bioinformatics 14:817–818
Ranjard L, Poly F, Nazaret S (2000) Monitoring complex bacterial community using culture-independent molecular techniques: application to soil environment. Res Microbiol 151:167–177
Rath J, Wu KY, Herndl GJ, DeLong EF (1998) High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol 14:261–269
Ravenschlag K, Sahm K, Pernthaler J, Amann R (1999) High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol 65:3982–3989
Satyanarayana T, Raghukumar C, Shivaji S (2005) Extremophilic microbes: diversity and perspectives. Curr Sci 89:78–90
Selenska-Pobell S, Kampf G, Flemming K, Radeva G, Satchanska G (2001) Bacterial diversity in soil samples from two uranium waste piles as determined by rep-APD, RISA and 16S rDNA retrieval. Antonie van Leeuwenhoek 79:149–161
Shima S, Yanagi M, Saiki H (1994) The phylogenetic position of Hydrogenobacter acidophilus based on 16S rRNA sequence analysis. FEMS Microbiol Lett 119:119–122
Skirnisdottir S, Hreggvidsson GO, Hjorleifsdottir S, Marteinsson VT, Petursdottir SK, Holst O, Kristjansson JK (2000) Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl Environ Microbiol 66:2835–2841
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland
Takacs CD, Ehringer M, Favre R, Cermola M, Eggertsson G, Palsdottir A, Reysenbach AL (2001) Phylogenetic characterization of the blue filamentous bacterial community from an Icelandic geothermal spring. FEMS Microbiol Ecol 35:123–128
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Whitaker RJ, Grogan DW, Taylor JW (2003) High-resolution multilocus sequence analysis revealed that, on a global scale, populations of hyperthermophilic microorganisms are isolated from one another by geographic barriers and have diverged over the course of their recent evolutionary history. Science 301:976–978
Yamamoto H, Hiraishi A, Kato K, Chiura HX, Maki Y, Shimizu A (1998) Phylogenetic evidence for the existence of novel thermophilic bacteria in hot spring sulfur-turf microbial mats in Japan. Appl Environ Microbiol 64:1680–1687
Acknowledgments
This work was supported by a grant from the Ministry of Education and Science of Bulgaria, Project B1511/05. We gratefully acknowledge the helpful technical advices of Dr. G. Radeva of the Institute of Molecular Biology, Bulgarian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tomova, I., Stoilova-Disheva, M., Lyutskanova, D. et al. Phylogenetic analysis of the bacterial community in a geothermal spring, Rupi Basin, Bulgaria. World J Microbiol Biotechnol 26, 2019–2028 (2010). https://doi.org/10.1007/s11274-010-0386-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-010-0386-7