Abstract
Forty-four Vibrio cholerae isolates collected over a 7-month period in Chennai, India in 2004 were characterized for gene traits, antimicrobial susceptibility and genomic fingerprints. All 44 isolates were identified as O1 El Tor Ogawa, positive for various toxigenic and pathogenic genes viz. ace, ctxB, hlyA, ompU, ompW, rfbO1, rtx, tcpA, toxR and zot. Nucleotide sequencing revealed the presence of cholera toxin B of classical biotype in all the El Tor isolates, suggesting infection of isolates by classical CTXΦ. Antibiogram analysis showed a broad-spectrum antibiotic resistance that was also confirmed by the presence of resistant genes in the genomes. All isolates contained a class 1 integron and an SXT constin. However, isolates were sensitive to chloramphenicol and tested negative for the chloramphenicol resistant gene suggesting a deletion in SXT constin. Fingerprinting analysis of isolates by ERIC- and Box PCR revealed similar DNA patterns indicating the clonal dissemination of a single predominant V. cholerae O1 strain throughout the 2004 outbreak in Chennai.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The disease cholera, caused by the bacteria Vibrio cholerae, is a continuous public health problem in countries with poor socio-economic conditions. Almost every developing country is facing either a cholera outbreak or the threat of an epidemic. Cholera has the potential to appear in explosive outbreak, epidemic or even pandemics. The world has already faced seven cholera pandemics in the past two centuries (Kaper et al. 1995). The Indian sub-continent has been an epicenter for cholera. Chennai, a metropolitan city in Southern India, is plagued by reoccurring cholera outbreaks in suburbs due to contamination of drinking water supplies. V. cholerae O139 strain, an epidemic strain that was unknown prior to 1991, emerged from this region and spread to the Southeast Asian countries.
The two biotypes of O1 serotype, El Tor and classical are believed to have evolved from separate lineages (Kaper et al. 1995). Between the two biotypes, El Tor strains have better adaptability to survive in the environment and in the human host as they colonize better the intestinal epithelium (Finkelstein 2006). Strains of classical biotype are suggested to be more toxigenic than El Tor strains (Huq et al. 1993). Recent studies have shown the spread of El Tor strains harbouring classical cholera toxin gene in US Gulf Coast and several countries in Asia and Africa (Nair et al. 2006; Goel et al. 2008).
In this study, we report molecular characterization of 44 V. cholerae O1 isolates from patients during a cholera outbreak in Chennai from April to November 2004. Strains were characterized for antimicrobial susceptibility, the presence of various toxigenic and pathogenic genes, nucleotide sequences of ctxB gene, and genomic fingerprints. This research contributes to our understanding of V. cholerae disease pattern and the evolution of pathogenicity.
Materials and methods
Bacterial cultures
The V. cholerae strains were isolated from stool samples of randomly selected patients during an outbreak in Chennai, India, between April and November 2004. Samples were collected using sterile rectal swabs from the patients admitted at Communicable Disease Hospital, Chennai as described earlier (Pourshafie et al. 2007). Forty-four isolates were randomly selected spanning the 7-month sampling period. V. cholerae was identified and confirmed by using standard biochemical methods (Tamrakar et al. 2006). Serological identification of the isolates was done by slide agglutination using commercially available polyvalent antiserum against V. cholerae O1 and O139 (Difco, USA). Biotype of isolates was confirmed by polymyxin B sensitivity and VP test. V. cholerae O1 (ATCC 11623) and V. cholerae O139 (ATCC 51394) were used as reference strains.
Biochemical characterization
All bacterial isolates were screened for oxidase reaction followed by other standard tests for presumptive identification of V. cholerae (Tamrakar et al. 2006). Serological identification of the isolates was done by slide agglutination using commercially available polyvalent antiserum against V. cholerae O1 and O139 (Difco, USA).
Antibiotic susceptibility
The antimicrobial susceptibility of the V. cholerae isolates was determined by the disc diffusion method on Mueller–Hinton agar as described by the Clinical Laboratory Standards Institute (CLSI 2007). The antibiotics discs were used at the following concentrations: ampicillin (10 μg), chloramphenicol (30 μg), ceftazidime (10 μg), ciprofloxacin (5 μg), co-trimoxazole (25 μg), gentamicin (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), nitrofurantoin (300 μg), norfloxacin (10 μg), polymyxin-B (50 μg), spectinomycin (100 μg), streptomycin (10 μg), sulphamethizole (300 μg), sulphamethoxazole (100 μg) tetracycline (30 μg) and trimethoprim (5 μg). The antibiotic discs used in this study were purchased from HiMedia, India.
Detection of gene traits
Genomic DNA was extracted from each of the isolate using genomic DNA purification kit (MBI Fermentas, Vilnius, Lithuania) and screened for the presence of diverse gene traits by two sets of multiplex PCR as described elsewhere (Kumar et al. 2009). The first multiplex PCR revealed the presence of ompW, ctxB, rfbO1, tcp and zot genes. The second set of multiplex PCR detected rtxC, ace, hlyA, ompU and toxR gens. Primer sequences used for amplification of different genes are listed in Table 1. The specificity of PCR was confirmed by using reference bacterial strains. PCR amplification was carried out in a 25 μl reaction mixture using a Palm Cycler (Corbett Life Sciences, Australia). The reaction mixture contained 1× reaction buffer, 200 μM dNTPs, 1.5 mM MgCl2, 1 U of Taq polymerase (Fermentas), varying concentration of primers (see Table 1) specific for each target gene, 100 ng of template DNA and milli-Q water. The thermal cycling conditions for both the multiplex PCR were: pre-incubation at 94°C for 2 min, 30 cycles of 1 min at 94°C for denaturation, 1 min at 59°C for annealing and 2 min at 72°C for extension followed by incubation at 72°C for 10 min for final extension.
PCR for class 1 integron and SXT constin genes
Class 1 integron was detected by PCR using primers qacEΔ1-F and sul1-B, directed at the 3′-CS of class 1 integrons. To investigate the presence of SXT constin, a conjugative self transmissible plasmid, primers int1-F and int1-B specific for SXT integrase were used (Hochhut et al. 2001). The primer sequences and PCR conditions used for class 1 integrons and SXT constin were identical to those described previously (Hochhut et al. 2001). To verify the presence of antibiotic resistant gene cassettes among integrons, primers in-F and aadA-B were used to detect the presence of aadA2 gene cassette encoding resistance to streptomycin and spectinomycin. Primers in-F and blaP1-B were used to detect the presence of β-lactam gene cassette in integrons (Dalsgaard et al. 2000). Isolates positive for SXT integrase gene were examined for the presence of the SXT-associated resistance genes, floR and strA (encoding for chloramphenicol and streptomycin resistance, respectively) by PCR as previously described (Hochhut et al. 2001).
Sequencing of cholera toxin (ctxB) gene
Cholera toxin B (ctxB) gene was amplified from the isolates using the ctxF and ctxR primers as described earlier (Olsvik et al. 1993). Sequencing was carried out using the same PCR primers on a 96 capillary model 3730xl system using the Big Dye Terminator kit from Applied Biosystems (Applied Biosystems, Foster City, CA, USA). The sequences were edited with SeqED program (Applied Biosystems), and were aligned with reference strains using ClustalW program. The sequences of ctxB gene for reference strains were retrieved from the public domain (GenBank).
Nucleotide sequence accession number
The nucleotide sequences obtained for the ctxB gene of strains VCC1588, VCC1969, VCC2411, VCC2491, VCC4372, VCC5910, VCC5963 and VCC5988 have been deposited in GenBank under accession number EU428013, EU428014, EU428015, EU428016, EU428017, EU428018, EU428019 and EU428020, respectively.
Genomic DNA fingerprinting analyses
Enterobacterial repetitive intergenic consensus (ERIC) sequence PCR was performed as described earlier with little modifications by using two oligonucleotides ERIC-F and ERIC-R (Rivera et al. 1995). The thermal cycler was programmed for 35 cycles of 1 min at 94°C, 1 min at 52°C, 10 min at 68°C followed by 20 min incubation at 70°C. BOX PCR was performed by using a single nucleotide primer BOX A1R (Versalovic et al. 1994). PCR program consisted of: initial denaturation (95°C for 7 min), 30 cycles of 94°C for 1 min, 53°C for 1 min, 65°C for 8 min, and a final extension of 65°C for 16 min.
Results and discussion
Strain identification and detection of gene traits
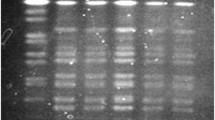
A total of 44 bacteria isolated from the patients were biochemically identified as V. cholerae and serologically confirmed as O1 Ogawa. Multiplex PCR analysis revealed that all strains tested positive for ace, ctxB, hlyA, ompU, ompW, rfbO1, rtx, tcpA, toxR and zot genes (Fig. 1). The outer membrane gene (ompW) is species specific for V. cholerae and its presence confirms the biochemical identification of V. cholerae. Somatic O-antigen biosynthesis gene was positive for O1 type (rfbO1), providing molecular evidence for O1 serogroup. Collectively, the results indicated that 2004 Chennai outbreak was caused by a toxigenic V. cholerae O1 Ogawa.
Multiplex PCR 1 (Lanes 1–5) and multiplex PCR 2 (Lanes 6–10) for analysis of various genes. Lanes 1 and 6: V. cholerae O1 (ATCC 11623); lanes 2 and 7: V. cholerae O139 (ATCC 51394); lanes 3 and 8: V. cholerae VCC1969; lanes 4 and 9: V. cholerae VCC2411; lanes 5 and 10: V. cholerae VCC5988; lane M: 100 bp ladder
To identify the biotype of the isolates, VP test, polymyxin B resistant test and rtxC PCR were performed. Results showed that all isolates were VP negative, resistant to polymyxin B and PCR positive for repeat in toxin (RTX) gene C (rtxC). Reference V. cholerae O1 classical (ATCC 11623) strain was PCR negative for RTX gene. Both negative VP test and polymyxin B resistance suggested that Chennai isolates belonged to El Tor biotype. The RTX represents a family of important virulence factors that have disseminated widely among gram-negative bacteria (Coote 1992). The RTX gene cluster in V. cholerae encodes the presumptive cytotoxin (rtxA), an acyltransferase (rtxC), and an associated ATP-binding cassette transporter system (Lin et al. 1999). PCR assays developed for rtxA or rtxC in V. cholerae can differentiate El Tor biotype from classical biotype among O1 serogroup (Chow et al. 2001). The positive amplification of rtxC gene among the isolates further confirmed El Tor biotype.
In addition to ctxB and rtxC genes, the isolates were also positive for haemolysin (hlyA), zonula occludens (zot), accessory cholera toxin (ace), toxin coregulated pilus (tcp), outer membrane protein (ompU) and toxR genes. Cholera pathogenesis is a complex process and involves synergetic action of several genes. Although cholera toxin (CT) is supposed to be the most important epidemic marker among various toxins produced by V. cholerae (Kaper et al. 1995), CT is encoded by a mobile element, the genome of a filamentous CTX bacteriophage. The acquisition of CTXϕ relies on the presence of toxin co-regulated pilus (encoded by tcp genes) on cell surface, which acts as a receptor for CTXϕ infection (Waldor and Mekalanos 1996). Zonula occludens toxin (zot) and accessory cholera toxin (ace) genes are, in fact, genes encoding for CTXϕ structural proteins (Waldor and Mekalanos 1996). The simultaneous detection of the suite of ancillary toxin genes with ctxB gene confirmed the presence of CTX prophage in Chennai isolates.
Alteration of ctxB gene among El Tor strains
Figure 2 shows the amino acid sequences deduced from the nucleic acid sequences of ctxB gene. Interestingly, the CTX B sequences from the Chennai outbreak strains aligned with O1 classical reference strains. The deduced amino acid sequences differed from that of the reference El Tor strain by a histidine at position 39 and a threonine at position 68. Thus, the results indicated that Chennai El Tor strains contained an altered cholera toxin gene. It is likely caused by infection by classical CTXΦ. More accurately, Chennai outbreak was caused by a “hybrid” El Tor strain with classical phage. Similar results have been reported earlier from other countries in Asia and Africa (Nair et al. 2006; Safa et al. 2008) and more recently in eastern India (Kumar et al. 2009). The discovery of El Tor strains with classical ctxB gene in Chennai outbreak implied a wide-spread of El Tor strains with the “newly acquired” classical toxin gene in the Indian subcontinent.
Amino acid sequence alignment of CTX-B subunit of V. cholerae O1 El Tor strains from Chennai outbreak with reference El Tor and classical strains. Identical amino acid residues are indicated by dots. Amino acid sequences of V. cholerae CTX-B reference strains used in alignment were retrieved from GenBank
The seventh and current pandemic of cholera is caused by the El Tor biotype. The classical biotype is believed to be extinct (Nair et al. 2002; Safa et al. 2008). The classical and El Tor biotypes of V. cholerae O1 are closely related in their O-antigen biosynthetic genes. However, the genomic structure of the CTXΦ, in which the cholera toxin genes are contained, differs between the classical and El Tor biotypes (Waldor and Mekalanos 1996; Ansaruzzaman et al. 2004). The results from India and Bangladesh showed that classical CT producing El Tor strains are now replacing the seventh pandemic El Tor strains (Nair et al. 2002, 2006; Kumar et al. 2009). This is taken as an evolutionary optimization of the El Tor biotype, which could represent a new and more significant emerging form of the El Tor biotype of V. cholerae.
Antibiotic resistant and mobile elements
Antibiotic susceptibility test revealed that all 44 isolates were sensitive to ampicillin, ceftazidime, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, norfloxacin and tetracycline. However, the isolates exhibited high rate of resistance towards co-trimoxazole, nalidixic acid, nitrofurantoin, spectinomycin, streptomycin, sulphamethizole, sulphamethoxazole and trimethoprim. All the isolates were resistant to polymyxin-B, a typical feature of El Tor biotype strains. Multiple antibiotic resistance among V. cholerae has emerged as a major problem worldwide (Faruque et al. 2007). In India, there is a progressive increasing trend of antibiotic resistance towards common fluoroquinolone, i.e., ciprofloxacin and norfloxacin since 1996 (Garg et al. 2001; Krishna et al. 2006). In this study, however, strains are sensitive to ciprofloxacin and norfloxacin but resistant to nalidixic acid, a non-fluorinated quinolone. In addition, isolates were sensitive to tetracycline, which is different from reports from other regions of India showing a re-emergence of tetracycling resistant strains (Jesudason 2006). Multi-drug resistance among V. cholerae strains limits the therapeutic potential of these drugs. Some antibiotics are already deemed unsuitable for certain group of population, i.e., tetracycline is not recommended for use in children and quinolone are not recommended in pregnant women and children (Sabeena et al. 2001). Multi-drug resistance presents additional challenges to disease management.
PCR results showed positive amplification of class 1 integron from Chennai isolates (Fig. 3). Integrons are an important mechanism for the acquisition of antibiotic resistance genes in many bacteria (Hall and Collis 1995). These elements are not autonomously mobile but are able to capture, integrate and express resistance gene cassettes in their variable region. In this study, isolates were found to have integrons on the basis of PCR with 3′CS conserved sequence. PCR results also confirmed the presence of aadA2 gene cassettes within the integron. The aadA2 gene cassettes encode aminoglycoside adenylyltransferases inactivating streptomycin and spectinomycin (Recchia and Hall 1995). These gene cassettes are among the most prevalent gene cassettes in class 1 and class 2 integrons. All isolates were negative for blaP1, the β-lactam gene cassette, corresponding to the phenotypic sensitivity of Chennai strains to ampicillin and ceftazidime.
Recent studies suggest SXT constin as an important element for horizontal dissemination of antibiotic resistant genes in bacteria (Beaber and Waldor 2004). In SXT constins, the antibiotic resistance genes are clustered within a composite transposon-like structure found near the 5′ end of SXT. These genes confer resistance to chloramphenicol, sulphamethoxazole, streptomycin and trimethoprim (Beaber et al. 2002). In this study, all strains were found PCR positive for SXT integrase gene suggesting the presence of STX constin (Fig. 3). They were resistant to sulphamethoxazole, streptomycin and trimethoprim but were sensitive to chloramphenicol. The presence of strA gene within SXT was confirmed by PCR, which mediates resistance to streptomycin by phosphotransferase enzymes (aminoglycoside-3′-phosphotransferase and aminoglycoside-6′-phosphotransferase (Shaw et al. 1993). However, all isolates were PCR negative for floR gene, which is responsible for resistance towards chloramphenicol (Fig. 3). The deletion of floR gene within SXT contin suggests evolution of resistance characteristics through mobile element. SXT variants have been reported among clinical V. cholerae isolates in recent years (Faruque et al. 2003; Iwanaga et al. 2004). Early reports indicated the loss of resistance to sulphamethoxazole and trimethoprim, and showed varying resistance to streptomycin (Mukhopadhyay et al. 1998; Faruque et al. 2003). The new SXT variant found in Chennai isolates reconfirmed the importance of mobile element in the development of antibiotic resistance. We have demonstrated a direct correlation between the antibiotic resistant phenotype and the presence of drug resistance genes in Class 1 integron and SXT constins among Chennai isolates.
DNA fingerprinting analyses
All 44 V. cholerae isolates were characterized by ERIC- and BOX PCR to reveal the clonal relationship. ERIC PCR with genomic DNA of V. cholerae strains resulted in amplification of multiple fragments of DNA ranging between 0.25 and 1.8 kb. The BOX PCR of genomic DNA resulted in fragments varying between 0.65 and 6.0 kb. The fingerprinting analysis revealed identical DNA banding pattern among all 44 isolates, suggesting that a single clone of V. cholerae likely caused the Chennai outbreak over the 7-month period.
In summary, the study demonstrated that V. cholerae O1 El Tor biotype was responsible for the seven-month Chennai outbreak in 2004. This El Tor strain, however, contained classical cholera toxin gene, suggesting infection of isolates by classical CTXΦ. Mobile elements in the strain conferred multiple antibiotic resistances. All the isolates were derived from a single clone that initiated the outbreak in the area. This study calls attention for continuous monitoring of outbreak strains to understand the evolution of pathogen and disease pattern.
References
Ansaruzzaman M, Bhuiyan NA, Nair BG, Sack DA, Lucas M, Deen JL, Ampuero J, Chaignat CL (2004) Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis 10:2057–2059
Beaber JW, Waldor MK (2004) Identification of operators and promoters that control SXT conjugative transfer. J Bacteriol 186:5945–5949
Beaber JW, Hochhut B, Waldor MK (2002) Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol 184:4259–4269
Chow KH, Ng TK, Yuen KY, Yam WC (2001) Detection of RTX toxin gene in Vibrio cholerae by PCR. J Clin Microbiol 39:2594–2597
CLSI (2007) Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement: CLSI document M100-S17 (ISBN 1-56238-625-5). Clinical and Laboratory Standard Institute, 940 West Valley Road, Suite 1400, Wayne Pennsylvania 19087-1898 USA
Collis CM, Hall RM (1992) Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol 174:1574–1585
Coote JG (1992) Structural and functional relationships among the RTX toxin determinants of gram-negative bacteria. FEMS Microbiol Rev 8:137–161
Dalsgaard A, Forslund A, Petersen A, Brown DJ, Dias F, Monteiro S, Molbak K, Aaby P, Rodrigues A, Sandstrom A (2000) Class 1 integron-borne, multiple-antibiotic resistance encoded by a 150-kilobase conjugative plasmid in epidemic Vibrio cholerae O1 strains isolated in Guinea-Bissau. J Clin Microbiol 38:3774–3779
Faruque SM, Sack DA, Sack RB, Colwell RR, Takeda Y, Nair GB (2003) Emergence and evolution of Vibrio cholerae O139. Proc Natl Acad Sci USA 100:1304–1309
Faruque AS, Alam K, Malek MA, Khan MG, Ahmed S, Saha D, Khan WA, Nair GB, Salam MA, Luby SP, Sack DA (2007) Emergence of multidrug-resistant strain of Vibrio cholerae O1 in Bangladesh and reversal of their susceptibility to tetracycline after two years. J Health Popul Nutr 25:241–243
Finkelstein RA (2006) Vibrio cholerae O1 and O139, and other pathogenic vibrios. Available online at http://gsbs.utmb.edu/microbook/ch024.htm
Garg P, Sinha S, Chakraborty R, Bhattacharya SK, Nair GB, Ramamurthy T, Takeda Y (2001) Emergence of fluoroquinolone-resistant strains of Vibrio cholerae O1 biotype El Tor among hospitalized patients with cholera in Calcutta, India. Antimicrob Agents Chemother 45:1605–1606
Goel AK, Ponmariappan S, Kamboj DV, Singh L (2007) Single multiplex polymerase chain reaction for environmental surveillance of toxigenic-pathogenic O1 and non-O1 Vibrio cholerae. Folia Microbiol (Praha) 52:81–85
Goel AK, Jain M, Kumar P, Bhadauria S, Kmboj DV, Singh L (2008) A new variant of Vibrio cholerae O1 El Tor causing cholera in India. J Infect 57:280–281
Hall RM, Collis CM (1995) Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol 15:593–600
Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK (2001) Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother 45:2991–3000
Huq A, Parveen S, Qadri F, Sack DA, Colwell RR (1993) Comparison of Vibrio cholerae serotype O1 strains isolated from patients and the aquatic environment. J Trop Med Hyg 96:86–92
Iwanaga M, Toma C, Miyazato T, Insisiengmay S, Nakasone N, Ehara M (2004) Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob Agents Chemother 48:2364–2369
Jesudason MV (2006) Change in serotype and appearance of tetracycline resistance in V. cholerae O1 in Vellore, South India. Indian J Med Microbiol 24:152–153
Kaper JB, Morris JG Jr, Levine MM (1995) Cholera. Clin Microbiol Rev 8:48–86
Kazama H, Kizu K, Iwasaki M, Hamashima H, Sasatsu M, Arai T (1995) Isolation and structure of a new integron that includes a streptomycin resistance gene from the R plasmid of Pseudomonas aeruginosa. FEMS Microbiol Lett 134:137–141
Krishna BV, Patil AB, Chandrasekhar MR (2006) Fluoroquinolone-resistant Vibrio cholerae isolated during a cholera outbreak in India. Trans R Soc Trop Med Hyg 100:224–226
Kumar P, Jain M, Goel AK, Bhadauria S, Sharma SK, Kamboj DV, Singh L, Ramamurthy T, Nair GB (2009) A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. J Med Microbiol 58:234–238
Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, Calia KE, Calderwood SB, Fraser C, Mekalanos JJ (1999) Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci USA 96:1071–1076
Mukhopadhyay AK, Basu I, Bhattacharya SK, Bhattacharya MK, Nair GB (1998) Emergence of fluoroquinolone resistance in strains of Vibrio cholerae isolated from hospitalized patients with acute diarrhea in Calcutta, India. Antimicrob Agents Chemother 42:206–207
Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA (2002) New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol 40:3296–3299
Nair GB, Qadri F, Holmgren J, Svennerholm AM, Safa A, Bhuiyan NA, Ahmad QS, Faruque SM, Faruque AS, Takeda Y, Sack DA (2006) Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol 44:4211–4213
Olsvik O, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth IK, Fields PI (1993) Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol 31:22–25
Pourshafie MR, Bakhshi B, Ranjbar R, Sedaghat M, Sadeghifard N, Zaemi Yazdi J, Parzadeh M, Raesi J (2007) Dissemination of a single Vibrio cholerae clone in cholera outbreaks during 2005 in Iran. J Med Microbiol 56:1615–1619
Recchia GD, Hall RM (1995) Plasmid evolution by acquisition of mobile gene cassettes: plasmid pIE723 contains the aadB gene cassette precisely inserted at a secondary site in the incQ plasmid RSF1010. Mol Microbiol 15:179–187
Rivera IG, Chowdhury MA, Huq A, Jacobs D, Martins MT, Colwell RR (1995) Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl Environ Microbiol 61:2898–2904
Sabeena F, Thirivikramji G, Radhakutty G, Indu P, Singh DV (2001) In vitro susceptibility of Vibrio cholerae O1 biotype El Tor strains associated with an outbreak of cholera in Kerala, Southern India. J Antimicrob Chemother 47:361–362
Safa A, Sultana J, Dac Cam P, Mwansa JC, Kong RY (2008) Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg Infect Dis 14:987–988
Shaw KJ, Rather PN, Hare RS, Miller GH (1993) Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163
Singh DV, Matte MH, Matte GR, Jiang S, Sabeena F, Shukla BN, Sanyal SC, Huq A, Colwell RR (2001) Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl Environ Microbiol 67:910–921
Tamrakar AK, Goel AK, Kamboj DV, Singh L (2006) Surveillance methodology for Vibrio cholerae in environmental samples. Int J Environ Health Res 16:305–312
Versalovic J, Schneider M, De Brulin FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5:25–40
Waldor MK, Mekalanos JJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914
Acknowledgments
The authors thank Director, Communicable Disease Hospital, Chennai, India for extending help in sample collection. Authors are thankful to Director, DRDE, Gwalior for providing necessary facilities and funds for the work. AKG is thankful to Department of Biotechnology, Ministry of Science and Technology, Government of India for providing DBT Overseas Associate Fellowship to conduct research at University of California, Irvine.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Goel, A.K., Jain, M., Kumar, P. et al. Molecular characterization of Vibrio cholerae outbreak strains with altered El Tor biotype from southern India. World J Microbiol Biotechnol 26, 281–287 (2010). https://doi.org/10.1007/s11274-009-0171-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-009-0171-7