Abstract
In this study, a reverse transcription loop-mediated isothermal amplification (RT-LAMP) was developed for detection of porcine epidemic diarrhea virus (PEDV). Six primers were designed to amplify the nucleocapsid (N) gene of PEDV. The optimization, sensitivity, and specificity of the RT-LAMP were investigated. The results showed that the optimal reaction condition for RT-LAMP amplifying PEDV N gene was achieved at 63°C for 50 min. The RT-LAMP assay was more sensitive than gel-based RT-PCR and enzyme-linked immunosorbent assay. It was capable of detecting PEDV from clinical samples and differentiating PEDV from Porcine transmissible gastroenteritis virus, Porcine rotavirus, Porcine pseudorabies virus, Porcine reproductive and respiratory syndrome virus, and Avian infectious bronchitis virus.
Similar content being viewed by others
Introduction
Porcine epidemic diarrhea (PED) is an infectious enteric disease characterized by acute enteritis and diarrhea in pigs, and the infection is more severe in neonates [1]. At present, PED has been a major concern in the swine industry, particularly, in the Asia and Europe, resulting in large economic losses [2–4]. The causative agent of PED is porcine epidemic diarrhea virus (PEDV), an enveloped and single-stranded RNA virus that belongs to the family Coronaviridae [5]. Coronavirus comprises three major viral structural proteins: spike (S, 180–220 kDa), membrane (M, 27–32 kDa), and nucleocapsid (N, 55–58 kDa) proteins [5, 6]. The S protein is a major viral antigen, binds to a cellular receptor for virus attachment to enter target cells and mediates viral attachment to target cells [7–13]. The M protein is a trans-membrane protein [14] and it is involved in the assembly process of viral nucleocapsid and membrane [15, 16]. The N protein of coronaviruses is a phosphorylated protein that interacts with virus genomic RNA, forming a helical ribonucleoprotein [17]. Therefore, it plays important roles in viral genome transcription, core formation and virus assembly [18]. Viral N protein is also conserved and can be used as a diagnostic target for detecting viral infection.
Loop-mediated isothermal amplification (LAMP) is a recently developed DNA amplification method [19]. LAMP uses four to six primers that recognize six to eight regions of target DNA, in conjunction with the enzyme Bst polymerase, which has strand-displacement activity. The synchronization DNA synthesis by these primers maintains the specificity of the method. Interestingly, the amplification step can be performed under isothermal conditions, resulting in the synthesis of a large amount of DNA. LAMP proceeds when the forward inner primer (FIP) anneals to the complementary region in the target DNA and initiates the first-strand synthesis. Next, the outer forward primer (F3) hybridists and displaces the first strand, forming a loop structure at one end. The resulting single-stranded DNA serves as a template for backward inner primer (BIP)-initiated DNA synthesis and subsequent outer backward (B3)-primed strand-displacement DNA synthesis. The formed dumbbell-shaped DNA stem loop structure serves as a template for subsequent hybridization between one inner primer and the loop, initiating the displacement DNA synthesis. The LAMP method may form the original stem loop and a new stem loop that is twice as long as the original one. The final products are stem loop DNAs with several inverted repeats of the target DNA and cauliflower-like structures bearing multiple loops [19].
At present, the LAMP approach has been applied for detecting of infectious pathogens. Examples include H5N1 avian influenza virus [20], hepatitis B virus [21], foot-and-mouth disease virus [22], etc. In this study, the authors developed a reverse transcription (RT)-LAMP using primers directed toward the N gene of PEDV. The convenience, sensitivity, and specificity of the established RT-LAMP indicate its advantages and utility in detecting PEDV.
Materials and methods
Cells and viruses
PEDV isolate HLJBY, Porcine transmissible gastroenteritis virus (TGEV), Porcine pseudorabies virus (PrV), Porcine rotavirus (PRV), Porcine reproductive and respiratory syndrome virus (PRRSV) and Avian infectious bronchitis virus (IBV) are propagated in susceptible cells.
Primers
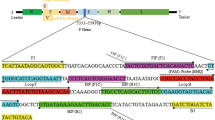
Based on the N gene sequence of PEDV (GenBank accession number: GU321197), a total of six primers targeting the N gene were designed using the Primer Explorer version 3 (http://primerexplorer.jp/lamp3.0.0/index.html). They include an outer pair (F3, B3), an inner pair (FIP, BIP), and a loop pair (F-loop, B-loop). A pair of primers (named PED1 and PED2) was used for RT-PCR amplifying the N gene. Information regarding the primer names and sequences is shown in Table 1.
PEDV propagation and RNA extraction
PEDV was propagated in African green monkey kidney (Vero) cells according to reference with modification [23]. In brief, Vero cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplementary with 10% newborn bovine serum (Excell Bio, China) in six-well plates at 37°C to allow the formation of cell monolayer. The cells were washed with PBS and infected with PEDV (500 μl/well) at an multiplicity of infection (MOI) of 2 at 37°C for 1 h in the presence of EDTA-free trypsin at a final concentration of 40 μg/ml. DMEM containing EDTA-free trypsin (40 μg/ml) was then added into the wells (2.5 ml/well) and the culture was maintained at 37°C for 48–36 h. The titer of PEDV was 106.25 TCID50/ml.
The total RNAs were extracted from the culture supernatants of PEDV, TGEV, PRV, PRRSV, and IBV using the RNA extraction kit (KeyGen Biotech, China) and the genomic DNA of PrV was extracted from virus-infected Vero cell culture using the DNA extraction kit (Omega, Norcross, USA) according to the manufacturer’s instructions. The extracted RNA was subjected to reverse transcription (RT) to synthesize the cDNA using reverse primer PED2 and a cDNA synthesis kit (HaiGene, China) according to the manufacturer’s instructions.
RT-LAMP reaction
The reaction mixture contained RNA template (2 μg) inner primers FIP and BIP (2 M/each), outer primers F3 and B3 (0.2 M/each), loop primers F-loop and L-loop (0.8 M/each), dNTPs mix (1.4 mM, HaiGene, China), 5 mM of MgSO4, Bst DNA polymerase (8 U, New England Biolabs, USA), 1× ThermoPol buffer consists of 20 mM Tris–HCL, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100 (HaiGene, China), and M-MuLV reverse transcriptase (HaiGene, China). The reaction was performed at a volume of 25 μl at 65°C in a water bath for 1 h and stopped at 85°C for 10 min. The sterile water was used as a negative control template. The amplified DNA products from the RT-LAMP were analyzed by separating 5 μl of RT-LAMP reaction mixture in ethidium bromide-stained 2% agarose gel electrophoresis, where the positive reaction mixtures showed a characteristic ladder of multiple bands. The relative quantification of the DNA was performed using the gel documentation system (Uvitec, Cambridge, UK) and determined with Gel Analyzer software (Copyright 2010 by Dr. Istvan Lazar) according to the manufacturer’s instructions. The reaction result was also observed directly without staining because of the white precipitate of magnesium pyrophosphate or the green color produced by the intercalating dye Picogreen® (Invitrogen, Wisconsin, USA) in positive reactions. After the amplification was completed, 2 μl of coloring agent (6× loading buffer:gene finder = 9:1) was added to each test tube and mixed. The test tubes were then examined visually.
Optimization of the RT-LAMP assay
To determine the optimal reaction temperature, the RT-LAMP reaction mixtures were incubated at 62, 63, 64, or 65°C for 30 min. The optimal reaction time was determined by performing the RT-LAMP at the optimal temperature for 10, 20, 30, 40, or 50 min. Finally, the reaction was terminated by heat inactivation at 80°C for 5 min. The amplified DNA products from the RT-LAMP assays were visualized by agarose gel electrophoresis as above.
Sensitivity analysis of the RT-LAMP
The concentration of PEDV RNA was determined using an ultra-violet photometer (Type 752, Shanghai Spectrum Instrument Company) according to the manufacturer’s instructions. Then the tenfold serial dilutions of the RNA (48 μg/ml) were used as template for RT-LAMP and a conventional RT-PCR. The RT-LAMP was performed as above. The RT-PCR was performed using a RT-PCR kit (HaiGene, China). The RT system included 1 μl of M-MuLV Reverse Transciptase (200 U/μl, TaKaRa, China), 4 μl of 5× RT-PCR buffer, 1 μl of dNTP Mixture (10 mM each), 2 μl of template, 1 μl of Oligo dT, 0.5 μl of RNase Inhibitor (40 U/μl), 11.5 μl of sterile water. The RT profile included 30°C for 10 min, 42°C for 60 min, and 95°C for 5 min. The PCR mixture was comprised of 12.5 μl of 2× HG PCR buffer, 2.5 μl of dNTP (2.5 mM), 0.5 μl of each primers (named PED1 and PED2, respectively), 1 μl of cDNA template, 0.5 μl of super Taq DNA polymerase (HaiGene), and 12. 5 μl of sterile water. The PCR parameters included 95°C for 5 min, 30 cycles of 94°C for 8 s, 47°C for 8 s, 72°C for 10 s, and a final extension at 72°C for 2 min.
ELISA was performed to compare its sensitivity with RT-LAMP. In brief, purified PEDV particles (0.1 μg/μl) were serially diluted in carbonate–bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) and the viruses were coated into ELISA plates (100 μl/well) at 4°C overnight. The next day, the plates were blocked with 5% non-fat dry milk in PBS-0.05% Tween 20 (PBST) at 37°C for 2 h. Subsequently, the wells were incubated with serially diluted anti-PEDV polyclonal antibody (1:1000 dilution) or control serum from a non-immunized rabbit at 37°C for 1 h, after triple wash with PBST. The plates were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (BOSTER, China 1:5000 dilution in PBST) at 37°C for 1 h. The wells were incubated with o-phenylenediamine dihydrochloride (OPD) substrate for 5 min after complete washing with PBST. The OD492 value was examined using an ELISA reader. The OD492 value of anti-PEDV serum positive well (P)/the OD492 value of control serum well (N) > 2 was regarded as positive.
Detection of PEDV in clinical samples by RT-LAMP, RT-PCR, and ELISA
Twenty clinical feces of piglets (approx. 3 weeks) with diarrhea symptom were collected from a pig farm in Heilongjiang Province in 2010. The samples were prepared as a 10% (w/v) suspension in PBS (pH 7.2) and centrifuged at 2000×g at 4°C for 10 min. The supernatant was subjected to RNA extraction with above-mentioned RNA extraction kit. The resulting RNA was used as a template for RT-PCR and RT-LAMP according to above-mentioned protocols. At the same time, the equal supernatant was used as coating antigen in ELISA as above. The detection limit of the three methods was compared.
Specificity of the RT-LAMP assay
To analyze the specificity of the RT-LAMP, PEDV, TGEV, PrV, PRV, PRRSV, and IBV were used as templates and subjected to RT-LAMP as above.
Results
RT-LAMP specific for detection of PEDV
Using PEDV RNA and six primers targeting the PEDV N gene, an RT-LAMP was done at 65°C in a water bath for 1 h. The resulting amplified DNA products showed a characteristic ladder of multiple bands, indicating that the final products were the mixtures of stem loop DNAs with various stem lengths (Fig. 1a). In contrast, the negative control did not show the characteristic bands. The results of the RT-LAMP reaction were also determined directly by visual inspection. If the reaction product is positive, the Gene Finder dye inserts into the double-stranded DNA after the reaction and the product becomes green; otherwise, the dye does not insert into the double-stranded DNA and the reaction sample remains blue (Fig. 1b).
RT-LAMP of PEDV N gene. PEDV RNA was used as template and six primers targeting the PEDV N gene was used in an RT-LAMP performed at 65°C for 1 h. The agarose electrophoresis results are shown in Panel a: Lane M DNA marker, lane 1 RT-LAMP product, lane 2 negative control. The reaction is also observed directly by adding of dye Picogreen® (Panel b). 1 Positive reaction, 2 negative reaction
Optimal temperature and time for the RT-LAMP assay
The effect of reaction temperature and incubation time on the RT-LAMP was investigated. As shown in Fig. 2a, the DNA products of the RT-LAMP at different temperatures showed multiple of characteristic ladder bands; however, the intensity of DNAs determined by Gel Analyzer software from the reactions at 63°C was stronger than that at other reaction temperatures, which was judged as the optimal temperature for RT-LAMP amplifying PEDV N gene. The RT-LAMP was then performed at 63°C for different time points. The results indicated that the DNA products showed the highest intensity when the reaction was performed for 50 min (Fig. 2b). Therefore, the optimal reaction condition of the current RT-LAMP for PEDV was 63°C for 50 min.
Optimization of the RT-LAMP of PEDV N gene. The optimization of the RT-LAMP targeting the PEDV N gene was analyzed by performing the reaction at 62, 63, 64, or 65°C for 30 min. The reaction results are shown in lanes 1–4, respectively (Panel a). The same reaction was performed at 63°C for 10, 20, 30, 40, or 50 min. The RT-LAMP results are shown in lanes 1–5, respectively (Panel b). Lane M DNA marker, lane 5 Negative control
Sensitivity comparison among RT-LAMP, RT-PCR, and ELISA
The sensitivity of the RT-LAMP assay was first compared with the conventional RT-PCR amplifying the tenfold serial dilutions of RNA templates of PEDV. The detection limit of RT-PCR was 4.8 × 10−2 μg/ml which equal to a virus titer of 102.25 TCID50/ml, while, the RT-LAMP had a detection limit of 4.8 × 10−6 (100.75 TCID50/ml) which was much higher than that of RT-PCR (Fig. 3). After applying the same concentration of PEDV particles in RT-LAMP and ELISA, the minimal required virus template amount for the both assays was analyzed. The results showed that the detection limit of RT-LAMP was 1 × 10−4 μg. In contrast, ELISA had a detection limit of 1 × 10−3 μg (Fig. 4).
Comparison of the sensitivity between RT-LAMP and RT-PCR. Tenfold serial dilutions of the PEDV RNA (48 μg/ml) were used as template for RT-LAMP and a conventional RT-PCR using specific primers. The results of RT-LAMP and RT-PCR are shown in panels a and b, respectively. Lane M DNA marker, lanes 1–8 The reaction results from serial dilution of RNA templates
Comparison of the sensitivity between RT-LAMP and ELISA. Purified PEDV particles (0.1 μg/μl) were serially diluted and used as coating antigen for an indirect ELISA or RT-LAMP. Lane M DNA marker, lanes 1–8 are the RT-LAMP results from 100 to 10−7 diluted virions. The OD492 value PEDV-containing positive well (P)/the OD492 value of control serum well (N) > 2 was regarded as positive reaction in ELISA. The P/N values of different virus dilution are indicated
Evaluation of clinical samples
RT-LAMP, RT-PCR, and ELISA were used simultaneously to detect 20 clinical samples. Their detection rates were 25% (5/20), 20% (4/20), and 25% (5/20), respectively (Table 2). The RT-LAMP had a similar sensitivity with ELISA and was somewhat sensitive than RT-PCR in detection of clinical samples.
The RT-LAMP distinguished PEDV from other viruses
To analyze the utility of the RT-LAMP, several related porcine viruses (i.e., TGEV, PRV, and PrV) and an avian coronavirus, IBV were used as templates and included in the RT-LAMP. The result indicated that no positive DNA products of the RT-LAMP assay were observed among these control viruses. When the PEDV was used as template, the positive bands were amplified as expected (Fig. 5). The result demonstrated that the RT-LAMP assay is specific and can be applied in discriminating ELISA for distinguishing PEDV from other viruses.
Discussion
There are PED epidemics in China, although inactivated vaccines are used in some regions in China. Establishment of rapid, sensitive, and cost-effective diagnostic assays for detecting PEDV is highly desirable. Virus isolation has been a popular detection method; nevertheless, the virological diagnosis is somewhat difficult for detecting PEDV, since it was not possible until 1988 to propagate porcine epidemic diarrhea virus in cell culture [23]. Even now, the viral titer of PEDV in cell culture is still low. Other diagnostic methods for detecting PEDV include immunohistochemistry, in situ hybridization, dot-blot hybridization, RT-PCR, and real-time RT-PCR [24–28]. These methods may require either high-precision instruments or complicated procedures. Therefore, they are unsuitable for detecting PEDV in fields and in less well-equipped laboratories.
The RT-LAMP method established in this study is a valuable alternative for detection of PEDV, since the novel DNA amplification technology owns numerous advantages such as simplicity, rapidity, and inexpensiveness. The isothermal conditions required for LAMP can be provided with a conventional water bath or heat block. Therefore, the current method can be applied less in well-equipped laboratories and fields for rapid detection of PEDV. In general, the LAMP can be carried out under isothermal conditions (60–65°C). In this study, the authors optimized the reaction conditions of the RT-LAMP by performing the test at different temperatures and time points. Subsequently, its sensitivity was compared with that of RT-PCR. The results showed that the RT-LAMP specific for PEDV was approx. 10,000 times sensitive than the RT-PCR. Nevertheless, it is necessary to screen other optimal primers to further compare the sensitivity between RT-LAMP and RT-PCR in the future. Moreover, the sensitivity between the RT-LAMP and conventional ELISA was compared using the inactivated PEDV as template. The former is more sensitive than the latter. Two reports have pointed out that the detection limit of RT-PCR for PEDV was 102.0 TCID50/ml [29, 30]. Detection limit of a commercially available ELISA kit (Jinma, Shanghai) used in China was 0.1 ng/ml, which was the same as the detection limit of the RT-LAMP developed in this study. This result further confirmed the sensitivity of the RT-LAMP for amplifying the N gene of PEDV. Nonetheless, when the authors used these methods to detect PEDV from clinical samples, the sensitivity of RT-LAMP was somewhat higher than RT-PCR and had a similar sensitivity with ELISA. More experiments are needed to clarify this point in the future; however, the RT-LAMP still has advantages including simplicity, rapidity, and convenience.
To analyze the specificity of the RT-LAMP for PEDV, several related or unrelated viruses were used as control templates. For example, PEDV and TGEV belong to the Group I coronaviruses which are closely related [31]. IBV and PRRSV belong to the Group III coronavirus and arterivirus, respectively; however, both viruses belong to the order Nidovirales [32, 33]. The structural similarity between the N proteins of IBV and PRRSV suggests that members of the Coronaviridae and Arteriviridae families share a mechanism of filamentous nucleocapsid formation, with suitable alterations necessary to interact specifically with their respective genomes [34, 35]. PrV and PRV are members of the families Herpesviridae and Reoviridae, respectively. These viruses such as PEDV, TGEV, PRV, PrV, or PRRSV may cause co-infection in pigs. The results showed that the RT-LAMP is successful only if the PEDV served as template, indicating that the established method is specific and applicable for differentiation diagnosis. To the knowledge, this is the first report regarding the establishment and optimization of a RT-LAMP for PEDV N gene. The assay may be useful for the clinical diagnosis of PEDV infection.
References
P. De Bouck, M.B. Pensaert, Am. J. Vet. Res. 41, 219–223 (1980)
G. Jimenez, J.M. Castro, M. del Pozzo, I. Correa, J. de la Torre, L. Enjuanes, in Proceedings of the International Pig Veterinary Society Congress, vol 9 (Mexico, 1986), p. 186
C. Chae, O. Kim, C. Choi, K. Min, W.S. Cho, J. Kim, J.H. Tae, Vet. Rec. 147, 606–608 (2000)
J.F. Chen, D.B. Sun, C.B. Wang, H.Y. Shi, X.C. Cui, S.W. Liu, H.J. Qiu, L. Feng, Virus Genes 36, 355–364 (2008)
F.A. Murphy, E.P.J. Gibbs, M.C. Horzinek, M.J. Studdert, Veterinary virology (Academic Press, San Diego, 1999)
H.F. Egberink, J. Ederveen, P. Callebaut, M.C. Horzinek, Am. J. Vet. Res. 49, 1320–1324 (1988)
M. Duarte, H. Laude, J. Gen. Virol. 75, 1195–1200 (1994)
C. Sune, G. Jimenez, I. Correa, M.J. Bullido, F. Gebauer, C. Smerdou, L. Enjuanes, Virology 177, 559–569 (1990)
I.M. Antón, S. González, M.J. Bullido, M. Corsín, C. Risco, J.P. Langeveld, L. Enjuanes, Virus Res. 46, 111–124 (1996)
X. Ren, J. Glende, M. Al-Falah, V. de Vries, C. Schwegmann-Wessels, X. Qu, L. Tan, T. Tschernig, H. Deng, H.Y. Naim, G. Herrler, J. Gen. Virol. 87, 1691–1695 (2006)
X. Ren, J. Glende, J. Yin, C. Schwegmann-Wessels, G. Herrler, Virus Res. 137, 220–224 (2008)
C. Schwegmann-wessels, J. Glende, X. Ren, X. Qu, H. Deng, L. Enjuanes, G. Herrler, J. Gen. Virol. 90, 1724–1729 (2009)
B. Liu, G. Li, X. Sui, J. Yin, H. Wang, X. Ren, J. Biotechnol. 141, 91–96 (2009)
A. Utiger, K. Tobler, A. Bridgen, M. Suter, M. Singh, M. Ackermann, Adv. Exp. Med. Biol. 380, 87–90 (1995)
P.J.M. Rottier, in The Coronaviridae, ed. by S.G. Siddel (Plenum Press, New York, 1995), pp. 115–139
C.A.M. de Haan, L. Kuo, P.S. Masters, H. Vennema, P.J.M. Rottier, J. Virol. 72, 6838–6850 (1998)
M.R. Macnaughton, M.H. Madge, J. Gen. Virol. 39, 497–504 (1978)
Y. Wang, X. Zhang, Virology 265, 96–109 (1999)
T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, T. Hase, Nucleic Acids Res. 28, e63 (2000)
M. Imai, A. Ninomiya, H. Minekawa, T. Notomi, T. Ishizaki, P.V. Tu, N.T.K. Tien, M. Tashiro, T. Odagiri, J. Virol. Methods 141, 173–180 (2007)
Q.Y. Li, Q.Y. Xu, N. Liu, F.X. Zhang, Lett. Biotech. 16, 647–648 (2005)
J.P. Dukes, D.P. King, S. Alexandersen, Arch. Virol. 151, 1093–1106 (2006)
M. Hofmann, R. Wyler, J. Clin. Microbiol. 26, 2235–2239 (1988)
K. Jung, J. Kim, O. Kim, B. Kim, C. Chae, J. Virol. Methods 108, 41–47 (2003)
K. Jung, C. Chae, J. Virol. Methods 123, 141–146 (2005)
O. Kim, C. Chae, Vet. Pathol. 37, 62–67 (2000)
O. Kim, C. Chae, Can. J. Vet. Res. 66, 112–116 (2002)
S. Escutenaire, N. Mohamed, M. Isaksson, P. Thorén, B. Klingeborn, S. Belák, M. Berg, J. Blomberg, Arch. Virol. 152, 41–58 (2007)
S.Y. Kim, D.S. Song, B.K. Park, J. Vet. Diagn. Invest. 13, 516–520 (2001)
D.S. Song, B.K. Kang, J.S. Oh, G.W. Ha, J.S. Yang, H.J. Moon, Y.S. Jang, B.K. Park, J. Vet. Diagn. Invest. 18, 278–281 (2006)
A. Bridgen, M. Duarte, K. Tobler, H. Laude, M. Ackermann, J. Gen. Virol. 74, 1795–1804 (1993)
D. Cavanagh, Arch. Virol. 142, 629–633 (1997)
X. Ren, M. Wang, J. Yin, G. Li, J. Biotechnol. 147, 130–135 (2010)
H. Jayaram, H. Fan, B.R. Bowman, A. Ooi, J. Jayaram, E.W. Collisson, J. Lescar, B.V. Prasad, J. Virol. 80, 6612–6620 (2006)
X. Ren, M. Wang, J. Yin, G. Li, J. Clin. Microbiol. 48, 1875–1881 (2010)
Acknowledgments
The authors acknowledge funding supported by Program for New Century Excellent Talents in Heilongjiang Provincial University (1155–NCET–005).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ren, X., Li, P. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of porcine epidemic diarrhea virus. Virus Genes 42, 229–235 (2011). https://doi.org/10.1007/s11262-011-0570-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-011-0570-3