Abstract
A unique feature of ascovirus infection is cleavage of host cells into virus containing vesicles. It has been suggested that the virus induces apoptosis, either by expression of a caspase or other means, which is then diverted toward vesicle formation. There is little known about the mechanism of vesicle formation. Recent genome sequences of three ascoviruses indicated the presence of several putative open reading frames coding for proteins that could be involved in lipid metabolism. These proteins may play a role in rearrangement of membranes in infected host cells leading to formation of vesicles. Here, we analyzed a lipase-like gene (ORF19) from Heliothis virescens ascovirus (HvAV-3e) expressed from 8 h after infection and essential for virus replication and cell cleavage. In addition, ORF19 knock down by RNA interference inhibited virus replication indicating that the gene is indispensable for HvAV-3e replication. However, under enzymatic assays tested, we did not detect any lipase or esterase activity from ORF19.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viruses in Ascoviridae are double stranded DNA, enveloped viruses with a large (110–186 kb) circular genome [1, 2]. They infect insects of the order Lepidoptera and are predominantly found in members of the family Noctuidae [2, 3]. The characteristic symptoms of ascovirus infection are stunted growth and loss of appetite [4, 5]. At the cellular level, ascoviruses produce a unique pathology. Infected cells invaginate to form vesicles that resemble apoptotic bodies, in which the virus assembles. Once the cell ruptures the vesicles disseminate through the hemolymph, giving the hemolymph a characteristic milky coloration [6–8]. Despite this uniqueness, little is known about the mechanisms involved in ascovirus replication and pathology.
To date, four ascovirus genomes have been sequenced: SfAV-1a [9], TnAV-2c [10], DpAV-4a [11], and HvAV-3e [12]. In the HvAV-3e genome, one of the open reading frames (ORFs), ORF19, was predicted to encode a 655 amino acid polypeptide with high homology to eukaryotic esterases and lipases. Of the other two ascovirus genomes published, an ortholog gene is present in SfAV-1a (ORF13, 47% identity) but not in TnAV-2c, although a homologue of the pancreatic lipase-like genes is present in TnAV-2c [10].
There are several examples of viruses encoding their own esterases/lipases as virulence factors. The vaccinia virus exists in two forms, as the highly infectious extracellular enveloped virion (EEV) and the less infectious intracellular mature virion (IMV). The virally encoded protein p37, which has been shown to have lipase activity [13], is essential for the formation of EEV from IMV and has been shown to play a role in viral membrane biogenesis and/or membrane fusion [14]. Also, a lipase homologue from Marek’s Disease Virus, has no lipase activity in vitro but was found to be an essential virulence factor in vivo [15]. It is clear that esterase/lipase-like homologues (with or without lipase activity) are important for normal replication and/or infectivity in a broad range of viruses. With this in mind, we sought to determine whether ORF19 has lipase/esterase activity and also if it is essential for HvAV-3e replication.
Materials and methods
Insects and cell lines
Spodoptera litura larvae were cultured at 25°C and 30% relative humidity. They were raised on an artificial diet consisting of wheat germ, yeast, soy flour, and agar as well as antibacterial (nipagin, sorbic acid, and Vitamin C) and antifungal agents (propionic acid and phosphoric acid). Helicoverpa zea fat body (Hz-FB; [16]) and Spodoptera frugiperda (Sf9) cell lines were cultured as monolayers at 27°C in SF-900II (Invitrogen) and Ex-Cell 420 (BD Biosciences) media, respectively.
Virus purification
This study was conducted on a strain of HvAV-3 isolated from South East Queensland, Australia (HvAV-3e). The virus was originally purified from infected Helicoverpa armigera larvae. The inoculum used to infect Hz-FB cells was amplified in Sf9 cells. Infected cells were collected 72 h post-infection, centrifuged at 2,000g and the supernatant collected as the inoculum.
For the in vivo inoculations, the milky-white hemolymph from infected (5 days) S. litura larvae was collected by anaesthetizing them on ice before removing one of the prolegs with fine-point forceps and squeezing the body of the caterpillar. The extract was stored at −80°C until used as the inoculum.
Construction and expression of recombinant baculovirus
To express ORF19 in Sf9 cells, the “Bac-to-Bac baculovirus expression system” (Invitrogen) was used. The full-length gene was amplified from purified viral genomic DNA (gDNA) by PCR using gene specific primers. The reverse primer also contained a poly-His (5′-GTGATG-3′ × 3) sequence following the last amino acid-encoding codon. The PCR product was cloned into pFastBac1 (Invitrogen) and sequenced in both directions to confirm the identity of the insert. The shuttle vector was then transferred into bacmid-containing E. coli DH10Bac cells by heat shock. Following confirmation, the recombinant bacmid was extracted and ~1 × 106 Sf9 cells were transfected with the bacmid using Cellfectin (Invitrogen) transfection reagent according to the “Bac-to-Bac” manual. The transfected cells were incubated at 27°C for 72 h to allow replication of the recombinant baculovirus. Expression of ORF19 was confirmed by western blotting using an anti-His antibody conjugated with alkaline phosphatase (1:5000; Sigma) and specific antibodies to ORF19 (1:5000). ORF19 was produced in Sf9 cells and purified for lipase assay using Ni–NTA beads (Qiagen). Extraction was confirmed by western blot analysis using anti-ORF19 polyclonal antibodies (1:5000).
Prediction studies
All predictions were carried out on software available from http://au.expasy.org. The molecular weight of ORF19 was predicted using Compute pI/MW. To determine the presence or absence of a signal peptide and where the protein localizes to, cell localization predictions were conducted using PSORT II. Glycosylation predictions were also undertaken using NetOGlyc v3.1, which predicts O-glycosylation sites and NetNGlyc v1.0, which predicts N-glycosylation sites.
Sequence alignment and 3D protein modeling
Sequence alignment studies were conducted using NCBI server (http://www.ncbi.nlm.nih.gov/) and formatted using Genedoc [17]. Tertiary structure prediction studies were also conducted using ESyPred3D v1.0 [18]. The model was generated using 1F8U chain A (human acetylcholinesterase mutant E202Q complexed with green mamba venom peptide) as a template, a protein with 27.5% identity to ORF19.
Temporal expression of ORF19
The in vitro temporal expression patterns of ORF19 were determined by reverse transcription polymerase chain reaction (RT–PCR). Hz-FB cell monolayers were incubated in 20 mm petri dishes at 27°C and infected with 200 μl of HvAV-3e inoculum described above. Cells from a single petri dish were resuspended, centrifuged (1 min at 16,000g), and the cell pellet frozen at −20°C at 2, 4, 8, 16, 24, 48, 72, and 96 h post-infection. Total RNA was extracted from cells using a previously described protocol [19]. All samples were then treated with DNase I (Promega) to remove possible DNA contamination.
Reverse transcription was performed at 42°C for 1 h with avian myeloblastosis virus reverse transcriptase (AMV-RT, Promega) using both ORF19-specific (5′-GACAATTTCACTGAGATAC-3′) and actin-specific (5′-GATGGCGTGGGGCAGGGC-3′, from Helicoverpa zea actin A3b cDNA AF286059) reverse primers. PCR was performed using ORF19-specific (Forward, 5′-ATCTCGACTGGCATACGC-3′) and actin-specific (Forward, 5′-ATGGAGAAGATCTGGCAC-3′) primers (reverse primers as above). Actin was used for normalizing the data as we found that its expression levels remained relatively unchanged following HvAV-3e infection (primers as above). Samples were also run without the addition of AMV-RT to ensure detection by PCR was not due to amplification of viral genomic DNA. The samples were run on 1% agarose gel containing ethidium bromide and viewed under UV light.
In addition, samples were subjected to quantitative RT–PCR using SYBR Green (Invitrogen) with a Rotor-Gene 6000. Primers used for qRT–PCR were orf19-For, 5′-ATCTCGACTGGCATACGC-3′; orf19-Rev, 5′-CGCAAAGTCCGTGAGTAGC-3′; Act-For, 5′-ATGGAGAAGATCTGGCAC-3′; and Act-Rev, 5′-GTTGGCCTTGGGGTTGAG-3′. After reverse transcription as above, real-time PCR conditions were 50°C for 2 min and 95°C for 2 min followed by 40 cycles of 95°C for 10 s, 60°C for 10 s, 72°C for 20 s, and final extension of 72°C for 20 s. Actin expression was used for normalizing the data. Relative DNA levels from each sample were compared in Qgene template program. Reactions were repeated three times.
Quantitative PCR
Total genomic DNA was extracted from cells and subjected to qPCR using specific primers to ORF19 from HvAV-3e genome. DNA concentrations were measured with Nanodrop and 50 ng total genomic DNA was used for each qPCR reaction using SYBR Green (Invitrogen) with a Rotor-Gene 6000. Real-Time PCR conditions were 50°C for 2 min and 95°C for 2 min followed by 40 cycles of 95°C for 10 s, 60°C for 10 s, 72°C for 20 s, and final extension of 72°C for 20 s. Actin was used for normalizing the data. Primers were as mentioned above in qRT–PCR. Relative DNA levels from each sample were compared in Qgene template program. Reactions were repeated three times.
Determination of localization of ORF19
To test localization in vitro, a culture of Sf9 cells was infected with recombinant baculovirus (as mentioned above) and incubated at 27°C for 72 h. The culture was then resuspended and centrifuged at 16,000g for 1 min, with both the cell pellet and the cell-free supernatant collected. The cell pellet, supernatant, and concentrated supernatant (10×) were mixed with 4× SDS–PAGE loading buffer and boiled for 5 min. The samples were then separated by SDS–PAGE (10%) and transferred to nitrocellulose for western blotting. The blot was probed with anti-ORF19 polyclonal antibodies (1:5000). The same process was performed on Hz-FB cells infected with HvAV-3e.
To test localization in vivo, 3rd instar S. litura larvae were infected with HvAV-3e and maintained at 23°C for 5 days. Approximately 50 μl of hemolymph was collected and separated into plasma and a cell pellet by centrifugation at 16,000g for 1 min. Fat bodies were also removed at this time for analysis. Each sample was separated by SDS–PAGE (10%) and analyzed by western blot using anti-ORF19 polyclonal antibodies.
Lipolytic assays
The concentration of ORF19 expression from the recombinant baculovirus was determined by Bradford assay using BSA as a calibrator (Sigma). A non-lipolytic wasp venom protein [20], Vn50, expressed in the same vector was also purified and the concentration calculated, to be used as a negative control. To test for lipase activity, the method of Colen et al. [21] was used. Briefly, 1.5 g of agar was added to 100 ml of Tris–Cl pH 8.8 along with 8 mg of Victoria blue B dye and 1.5 ml of a 20% olive oil in polyvinyl alcohol (2% PVOH in water) emulsion. The mixture was autoclaved and then used to pour a 35 mm plate. Wells were cut using a rivet sterilized in 100% ethanol. Three different concentrations of Candida rugosa lipase (Sigma) were used as positive controls. The concentration of the samples was adjusted to fall between those of the controls. Each well was loaded and the plate incubated at 30°C for ~17 h.
Esterase activity was tested using a similar method. The substrate was prepared as above except that instead of an olive oil in PVOH emulsion, 1 ml of tributyrin (glyceryl tributyrate, Sigma) was added to the mixture. Wells were cut and loaded as above. The plate was then incubated at 30°C for 3.5 h.
RNAi silencing
First, DNA fragments ~500 bp in size were amplified by PCR from both ORF19 and the jellyfish protein, green fluorescent protein (GFP) using forward and reverse primers containing T7 promoter sequences at their 5′ end (TAATACGACTCACTATAGGG). Double stranded RNA (dsRNA) was then produced and purified (by LiCl precipitation) for each fragment using the MEGAScript kit (Ambion) according to the manufacturer’s instructions. The concentration of RNA was determined by measuring absorbance at 260 nm.
To induce RNA silencing in vitro, Hz-FB cells were resuspended and ~1 × 103 cells added to individual wells of a 12-well plate. Once the monolayers had reformed (~1 h) the medium was removed and a transfection medium was added. This medium consisted of 0.5 ml of SF-900II, 5 μl of Cellfectin (Invitrogen), and 1 μl dsRNA, where dsRNA had been diluted to give final concentrations of 10, 5, or 1 nM for ORF19 and 10 nM for GFP, respectively. Each treatment was replicated three times. Following incubation for 5 h at 27°C, the transfection mixture was removed and 0.5 ml of fresh SF-900II medium (containing penicillin and streptomycin) was added. Twelve hours after removal of the transfection medium, each well was infected with 200 μl of HvAV-3e inoculum. The plate was then incubated at 27°C and observed daily by light microscopy. After 72 h post-infection (p.i.) all samples were resuspended and centrifuged. The supernatant was removed and the cell pellets were resuspended in SDS–PAGE loading buffer. The cellular proteins were then separated by SDS–PAGE (10%) and analyzed by western blot using polyclonal anti-capsid protein antibodies (1:5000). Cells were also subjected to qPCR as above.
Confocal microscopy
To analyze temporal localization of ORF19, Sf9 cells at 24, 48, and 72 h p.i. with HvAV-3e and non-infected cells were prepared for confocal microscopy as described previously [22]. Anti-ORF19 was used as the primary antibody (1:500) and an anti-rabbit antibody conjugated with FITC (1:100) was used as the secondary antibody.
Results
Sequence and characterization of HvAV-3e ORF19, a lipase homologue
The predicted molecular weight of the polypeptide encoded by ORF19 is 73.3 kDa. Many previously characterized lipases are secreted proteins, however, our localization prediction studies using prediction software suggested that ORF19 does not contain a signal/anchor peptide and is not targeted to secretory pathways, which is consistent with localization results below. In addition, an endoplasmic reticulum membrane retention signal at the N-terminus (XXSR) was found in the sequence (CKSR) indicating that the protein is not secreted. Glycosylation predictions detected three putative N-glycosylation sites as well as weakly detecting one putative O-glycosylation site.
Sequence alignment analyses showed that ORF19 has two substitutions in the conserved GXSXG motif found in lipases. First, there is an unfavorable substitution at the catalytic serine residue for a glycine and second, there is a substitution of the carboxyl-end glycine for a serine (GXSXG => GXGXS; Fig. 1). Glycine is an amino acid without a hydroxyl in its side group. This suggests that ORF19 is probably unable to perform the same catalytic activity as its homologues. In addition, protein modeling using ESyPred3D suggested that the three residues of the “catalytic triad” in ORF19 take on the same configuration as those in catalytically active lipases, the only difference being that the glycine in ORF19 does not have a hydroxyl group facing the histidine (Fig. 2). This finding further indicates that ORF19 is unlikely to have lipolytic activity.
Amino acid sequence alignment of ORF19 from HvAV-3e with SfAV-1a homologue and other eukaryotic esterases/lipases. Catalytic triad residues are indicated by ‘#’. Residues conserved in all sequences are shown in black; residues conserved in at least four sequences are shown in gray. HvAV_lip = ORF19; SfAV_lip = Spodoptera frugiperda ascovirus 1a ORF13; CR_lip = Candida rugosa lipase; CC_chl_est = Candida cylindracea cholesterol esterase; H_vir_JHE = juvenile hormone esterase precursor from Heliothis virescens; S_non_est = Sesamia nonagrioides esterase; and Hmn_BA_lip = human bile salt-activated lipase
Temporal expression pattern of ORF19
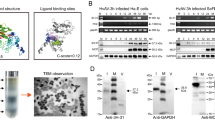
Expression of ORF19 was detected by RT–PCR from 8 h p.i. (very weakly) up until 96 h p.i. in the Hz-FB cell line (Fig. 3a). When the RT–PCR protocol was performed without the addition of reverse transcriptase ORF19 was not detected (data not shown). Since conventional RT–PCR is not quantitative and there was a slight reduction in the transcript levels at 72 hpi, we carried out RT–qPCR analysis of the samples which showed that expression of the gene gradually increased from 24 h p.i. and transcripts continued to accumulate up to 96 h p.i. (Fig. 3b).
Expression of ORF19. Temporal expression of ORF19 in Hz-FB cells infected with HvAV-3e analyzed by: a RT–PCR showing expression of ORF19 (top row) and actin (bottom row). M = mock infected sample; MW = GeneRuler 100 bp DNA molecular weight marker; and b RT–qPCR. Error bars represent standard deviations of averages from three replicates. c Western blot analysis showing temporal expression of recombinant ORF19 expressed by baculovirus. Cells infected with recombinant viruses expressing an unrelated protein (Vn50) were used as control. d Western blot analysis of cells expressing recombinant ORF19 (lane 2) and medium collected from cells (lane 3) and concentrated 10× (lane 4). ORF19 expressed in bacteria (lane 5) and Sf9 cells expressing Vn50 (lane 6) were used as positive and negative controls, respectively. All blots were probed with anti-ORF19 polyclonal antibodies (1:5000)
When expressed by the recombinant baculovirus, ORF19 was detected by western blot as a band ~74 kDa in size at 72 and 96 h p.i. in Sf9 cells. The predicated size for the protein including the His-tag is 75 kDa which is close to the size of the expressed protein. No bands were observed in uninfected cells, or cells infected with a recombinant baculovirus expressing a venom protein (Fig. 3c). Moreover, when the culture suspension was separated by centrifugation and both components analyzed by western blot, ORF19 was only detected in the cell pellet with a very strong band at ~74 kDa (Fig. 3d, lane 2). ORF19 was not detected from the cell-free medium, even when concentrated 10 times indicating that the protein is not secreted from cells (Fig. 3d, lanes 3 and 4). In addition, the protein was not found to be glycosylated when ECL glycosylation detection module (GE Healthcare) was applied (data not shown).
Lipolytic activity
To find out whether ORF19 has lipolytic activity, the purified recombinant protein was used in enzyme assays. At a concentration of 16 μg/ml, ORF19 had no detectable lipase activity using olive oil as a substrate. In contrast, lipase activity was detected in a lipase from Candida rugosa at a concentration of 6, 12, and 12 mg/ml (Fig. 4a; left panel). When assayed for esterase activity using tributyrin as a substrate, ORF19 again showed no enzymatic activity after 3.5 h whereas the lipase from C. rugosa at similar concentrations showed activity after 30 min (Fig. 4a; right panel).
a Assays for lipase (Lip) and esterase activities (Est). Blue halo indicates lipid hydrolysis. (Lip): Top left: ORF19 (664 ng); Top right: Candida rugosa lipase (480 μg); Center: C. rugosa lipase (480 ng); Bottom left: C. rugosa lipase (240 ng); Bottom right: Vn50 (negative control; 528 ng). (Est): Top left: C. rugosa lipase (240 μg); Top right: ORF19 (332 ng); Center: C. rugosa lipase (120 ng); Bottom left: Vn50 (264 ng); Bottom right: C. rugosa lipase (240 ng). b Western blot analysis of Hz-FB cells 72 hpi following transfection with ORF19 (at three concentrations) and GFP dsRNAs. The blot was probed with anti-AV-capsid polyclonal antibodies. Mock = no dsRNA added; MCP = major capsid protein. c RNAi in Hz-FB cells at 72 h p.i. showing: (A) uninfected cells; (B) cells infected with HvAV-3e only; (C) uninfected cells transfected with ORF19 dsRNA (10 nM); (D) cells infected with HvAV-3e following transfection with ORF19 dsRNA (10 nM); and (E) cells infected with HvAV-3e following transfection with GFP dsRNA (10 nM). d qPCR analysis of cells treated as in (b) with two concentrations of ORF19 and GFP dsRNA (5 and 10 nM). Control cells were mock transfected and infected with AV (M + AV). Error bars represent standard deviations of averages from three replicates. (Color figure online)
ORF19 is essential for virus replication and pathology
In vitro RNAi studies showed that ORF19 is crucial for normal viral replication. Transfection of Hz-FB cells with ORF19 specific dsRNA, to a final concentration of 10 nM, completely inhibited virus replication as no ascovirus capsid protein was detected in the cells 72 h following inoculation, as determined by western blot with polyclonal anti-capsid protein antibodies (Fig. 4b). ORF19 dsRNA at a final concentration of 5 and 1 nM each showed reduced detection of the capsid protein. The uninfected cells showed no capsid protein detection and cells that were transfected with GFP dsRNA added to a final concentration of 10 nM showed capsid protein levels comparable to that of cells mock-transfected or with 1 nM ORF19 dsRNA (Fig. 4b). These results were also confirmed by qPCR demonstrating that silencing of ORF19 significantly reduced viral DNA replication compared to mock or GFP dsRNA transfections (Fig. 4d).
When the above samples were examined by light microscopy there were several interesting observations (Fig. 4c). First, the cells infected with HvAV-3e and co-transfected with ORF19 dsRNA to a final concentration of 10 nM showed a marked reduction in the deep invaginations of the plasmalemma, vesiculation, and other malformations of the cell characteristics of HvAV-3e infection (Fig. 4c; D). Also, cells that were transfected with ORF19 dsRNA but left uninfected were not distinguishable from untreated cells (Fig. 4c; C). Control cells transfected with 10 nM GFP dsRNA showed pathology comparable to AV-only infected cells (Fig. 4c; E).
Localization of ORF19
To analyze temporal localization of ORF19, Sf9 cells at 24, 48, and 72 h p.i. with HvAV-3e were prepared for confocal microscopy. Results indicated that the protein is localized to the cytoplasm in foci that are most likely associated with endosomes (Fig. 5). As infection progressed, cells increased in size and the strength of the signal and the number of foci increased. In non-infected cells used as control mostly background staining was observed (Fig. 5).
Discussion
When insect cultured cells (Sf9 or Hz-FB) are infected with HvAV-3e they start to detach from the substrate and show cell enlargement by 24 h p.i. Major capsid protein gene expression starts from about 16 h p.i. followed by accumulation of the protein [23, 24]. By 48–72 h p.i. large invaginations of the plasmalemma, blebbing, and vesiculation of cells are observed [23, 25]. Similar observations were made in Spodoptera exigua AV (SeAV-5a = HvAV) [8] and Trichoplusia ni AV (TnAV-2a) [26]. Here, we showed that the HvAV-3e ORF19 is expressed from 8 h up until at least 4 days p.i. in vitro. The fact that it is expressed by 8 h p.i. is indicative that the protein is important during the initial phase of infection as well as during virion maturation at 2–4 days p.i. Our sequence alignment searches with known esterases/lipases showed that even though ORF19 has several conserved blocks of residues surrounding the three catalytic triad residues, there is a non-conserved substitution at the serine nucleophile motif, a motif conserved among all known active esterases and lipases. In ORF19 the catalytic serine has been substituted for a glycine. This same substitution is also present in the ortholog from SfAV-1a.
Only two lipolytic substrates were tested in this study but they are typical substrates for these type of enzymes. The assay using olive oil was less sensitive than that for esterase, requiring an overnight incubation to develop a distinct blue ring around the sample wells. In contrast, the esterase assay took only 15 min for color development to begin and the assay was stopped within 4 h of beginning. To confirm that there really was no activity by ORF19, rather than just a low level of activity, the esterase assay was also performed overnight and still no color development was observed. The substitution at the active site correlates with the finding that neither esterase nor lipase activities were detected, suggesting that its function may not be in lipolysis.
Prediction studies suggested the absence of a putative secretory signal peptide. This was consistent with our experimental results in which we were only able to detect ORF19 intracellularly and no glycosylation was detected on the protein. If ORF19 does contain an N-terminal transmembrane region yet remains uncleaved, then it will remain imbedded in the membrane. Confocal microscopy results seem to support this view. If this turns out to be the case, then it is possible that the protein may actually be targeted to other intracellular membranes, or that secretory vesicles are hijacked by viral processes and internalized. Alternatively, this protein might be a virion envelope protein involved in cell infection.
The study also demonstrated that ORF19 is essential for viral replication, since knock-down of the gene by RNAi abolished ascovirus pathology and reduced capsid protein production in vitro. This finding has similarities to the lipase homologue from Marek’s Disease Virus (MDV), where the MDV protein vLIP was found to have no lipolytic activity yet was still essential for disease development [15]. Similarly, there is a substitution in the catalytic triad of vLIP; the serine is substituted for a glycine. No specific function for vLIP in MDV biology has been determined. The exact mechanism(s) by which ORF19 influences replication is/are still unknown and requires further investigation. Considering this, when ORF19 was expressed independently from HvAV-3e, as in the recombinant baculovirus, there were no distinct pathological or morphological effects on Sf9 cells. Therefore, it can be postulated that ORF19 may be dependent on other viral components for normal function, rather than acting independently. Considering our findings that: (1) ORF19 remains inside the cytoplasm, (2) no esterase or lipase activities were detected, and (3) knock-down of the gene by RNAi prevented normal cell vesiculation as well as inhibiting viral replication, it is possible that ORF19 is targeted to intracellular membrane structures and plays a role in lipid-binding and/or in coalescing lipids into the replication vesicles.
References
B.A. Federici, J.M. Vlak, J.J. Hamm, Comparative study of virion structure, protein composition and genomic DNA of three ascovirus isolates. J. Gen. Virol. 71, 1661–1668 (1990)
X.-W. Cheng, G.R. Carner, T.M. Brown, Circular configuration of the genome of ascoviruses. J. Gen. Virol. 80, 1537–1540 (1999)
J.J. Hamm, D.A. Nordlung, O.G. Marti, Effects of a nonoccluded virus of Spodoptera frugiperda (Lepidoptera: Noctuidae) on the development of a parasitoid, Cotesia marginiventris (Hymenoptera: Braconidae). Environ. Entomol. 14, 258–261 (1985)
B.A. Federici, Enveloped double-stranded DNA insect virus with novel structure and cytopathology. Proc. Natl Acad. Sci. USA 80, 7664–7668 (1983)
R. Govindarajan, B.A. Federici, Ascovirus infectivity and effects of infection on the growth and development of noctuid larvae. J. Invertebr. Pathol. 56, 291–299 (1990)
B.A. Federici, R. Govindarajan, Comparative histopathology of three ascovirus isolates in larval noctuids. J. Invertebr. Pathol. 56, 300–311 (1990)
J.J. Hamm, E.L. Styer, B.A. Federici, Comparison of field-collected ascovirus isolates by DNA hybridization, host range and histopathology. J. Invertebr. Pathol. 72, 138–146 (1998)
X.-W. Cheng, G.R. Carner, B.M. Arif, A new ascovirus from Spodoptera exigua and its relatedness to the isolate from Spodoptera frugiperda. J. Gen. Virol. 81, 3083–3092 (2000)
D.K. Bideshi, M.V. Demattei, F. Rouleux-Bonnin, K. Stasiak, Y. Tan, S. Bigot, Y. Bigot, B.A. Federici, Genomic sequence of the Spodoptera frugiperda ascovirus 1a, an enveloped, double stranded DNA insect virus that manipulates apoptosis for viral reproduction. J. Virol. 80, 11791–11805 (2006)
L. Wang, J. Xue, C.P. Seaborn, B.M. Arif, X.-W. Cheng, Sequence and organization of the Trichoplusia ni ascovirus 2C (Ascoviridae) genome. Virology 354, 167–177 (2006)
Y. Bigot, S. Renault, J. Nicolas, C. Moundras, M.V. Demattei, S. Samain, D.K. Bideshi, B.A. Federici, Symbiotic virus at the evolutionary intersection of three types of large DNA viruses; Iridoviruses, Ascoviruses, and Ichnoviruses. PLoS One 4, e6397 (2009)
S. Asgari, J. Davis, D. Wood, P. Wilson, A. McGrath, Sequence and organization of Heliothis virescens ascovirus genome. J. Gen. Virol. 88, 1120–1132 (2007)
S.H. Baek, J.Y. Kwak, S.H. Lee, T. Lee, S.H. Ryu, D.J. Uhlinger, J.D. Lambeth, Lipase activities of p37, the major envelope protein of vaccinia virus. J. Biol. Chem. 272, 32042–32049 (1997)
R. Blasco, B. Moss, Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J. Virol. 65, 5910–5920 (1991)
J.P. Kamil, B.K. Tischer, S. Trapp, V.K. Nair, N. Osterrieder, H.J. Kung, vLIP, a viral lipase homologue, is a virulence factor of Marek’s disease virus. J. Virol. 79, 6984–6996 (2005)
A.H. McIntosh, C.M. Ignoffo, Replication and infectivity of the single-embedded nuclear polyhedrosis virus, Baculovirus heliothis, in homologous cell lines. J. Invertebr. Pathol. 37, 258–264 (1981)
P. Horton, K. Nakai, Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 147–152 (1997)
J.D. Bendtsen, H. Nielsen, G. von Heijne, S. Brunak, Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004)
K. Julenius, A. Molgaard, R. Gupta, S. Brunak, Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15, 153–164 (2005)
G.M. Zhang, Z.Q. Lu, H.B. Jiang, S. Asgari, Negative regulation of prophenoloxidase (proPO) activation by a clip-domain serine proteinase homolog (SPH) from endoparasitoid venom. Insect Biochem. Mol. Biol. 34, 477–483 (2004)
G. Colen, R.G. Junqueira, T. Moraes-Santos, Isolation and screening of alkaline lipase-producing fungi from Brazilian savanna soil. World J. Microbiol. Biotech. 22, 881–885 (2006)
S. Asgari, O. Schmidt, A coiled-coil region of an insect immune suppressor protein is involved in binding and uptake by hemocytes. Insect Biochem. Mol. Biol. 32, 497–504 (2002)
S. Asgari, Replication of Heliothis virescens ascovirus in insect cell lines. Arch. Virol. 151, 1689–1699 (2006)
M. Hussain, R.J. Taft, S. Asgari, An insect virus-encoded microRNA regulates viral replication. J. Virol. 82, 9164–9170 (2008)
S. Asgari, A caspase-like gene from Heliothis virescens ascovirus (HvAV-3e) is not involved in apoptosis but is essential for virus replication. Virus Res. 128, 99–105 (2007)
K. Zhao, L. Cui, Molecular characterization of the major virion protein gene from the Trichoplusia ni ascovirus. Virus Genes 27, 93–102 (2003)
Acknowledgment
This project was funded by a UQ internal grant to S. Asgari and a UQ PhD scholarship to Hussain.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Smede, M., Hussain, M. & Asgari, S. A lipase-like gene from Heliothis virescens ascovirus (HvAV-3e) is essential for virus replication and cell cleavage. Virus Genes 39, 409–417 (2009). https://doi.org/10.1007/s11262-009-0407-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-009-0407-5