Abstract
To investigate the prevalence of influenza viruses in feral water birds in the Southern Hemisphere, fecal samples of terns were collected on Heron Island, Australia, in December 2004. Six H2N5 influenza viruses were isolated. This is the first report of the isolation of the H2 subtype from shore birds in Australia. Phylogenetic analysis revealed that the M gene belonged to the American lineage of avian influenza viruses and the other genes belonged to the Eurasian lineages, indicating that genetic reassortment occurs between viruses of Eurasian and American lineages in free flying birds in nature.
Similar content being viewed by others
Introduction
Influenza viruses have two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), and are divided into H1 ~ 16 and N1 ~ 9 subtypes based on their antigenic specificity [1–5]. All subtypes of nonpathogenic influenza viruses have been isolated from feral water birds such as the Anseriformes (particularly ducks, geese, and swans) and Charadriiformes (particularly gulls, terns, and waders) [6]. Many Anseriformes and Charadriiformes are known to perform regular long-distance migrations [7], thereby potentially distributing nonpathogenic viruses between countries or even continents.
Since 2003, H5N1 highly pathogenic avian influenza (HPAI) viruses related to the viruses in Hong Kong during 1997 [8] have emerged and now appear to be endemic in many countries through southeast Asia, the Far east, and Africa [8–10]. A/chicken/Yamaguchi/7/04 (H5N1) isolated in Japan grows in multiple organs of ducks lacking overt signs and sheds from the infected ducks for approximately one week [11], although some H5N1 strains cause significant disease in several species of waterfowl [11–13]. If ducks infected with H5N1 do not show any signs of disease, as is the case with infections of A/chicken/Yamaguchi/7/04 (H5N1), it would be difficult to detect infections early on, paving the way for the transmission of HPAI viruses. Therefore, it cannot be ruled out that migratory birds transmit HPAI viruses.
To provide information on the precursor genes of future pandemic influenza viruses, we have been conducting virological surveillance of avian influenza since 1977 in the Northern hemisphere, namely, Alaska, Siberia, Russia, Mongolia, China, Taiwan, and Japan [5, 14–17]. However, information on the epidemiology of influenza in feral birds in the Southern hemisphere is not readily available. Therefore, in the present study, surveillance was carried out in Australia to investigate the prevalence of influenza viruses in feral water birds. Fecal samples were collected from terns on Herron Island in December 2004 and six H2N5 strains were isolated. These viruses were phylogenetically analyzed.
Materials and methods
Isolation and identification of viruses
We collected 1,413 fecal pellets dropped by terns on vinyl sheets spread under the trees where they sat at 30 different points on Heron Island, Australia, every morning and afternoon for 5 days in December 2004. The fecal samples were kept cool at 4°C until assayed. Each sample was suspended in phosphate buffer saline containing antibiotics and centrifuged. The supernatant was inoculated into the allantoic cavities of two 10-day-old chicken embryos and incubated for 48 h at 35°C. The allantoic fluid was collected and tested for hemagglutinating activity. The subtype of each virus isolate was determined with standard hemagglutination-inhibition (HI) and neuraminidase-inhibition (NI) tests using specific antisera to reference strains of influenza virus [17].
Viruses
In addition to the isolates from the fecal samples of terns in Australia, we used A/duck/Hong Kong/174/77 (H4N5), A/duck/Hong Kong/668/79 (H4N5), A/duck/Hokkaido/1058/01 (H4N5), A/duck/Hong Kong/943/80 (H6N5), A/chicken/Taiwan/4801/90 (H6N5), A/duck/Hong Kong/934/80 (H10N5), A/duck/Mongolia/149/03 (H10N5), A/duck/Hokkaido/24/04 (H10N5), A/duck/Hong Kong/862/80 (H12N5), A/duck/Hokkaido/66/01 (H12N5), and A/duck/Hokkaido/88/01 (H12N5) from our repository as reference strains of N5 for phylogenetic analysis.
Extraction of RNA and RT-PCR
Viral RNA was extracted from infectious allantoic fluids with Trizol LS reagent (Invitrogen, USA) and reverse-transcribed using oligonucleotide universal primers of influenza virus genes, Uni 12: 5′-AGC AAA AGC AGG-3′ and M-MLV Reverse Transcriptase (Invitrogen, USA). The RT-reaction was performed at 42°C for 60 min. The cDNA was amplified by polymerase chain reaction (PCR) with GoTaq® DNA Polymerase (Promega, USA) according to the protocol provided. The amplification program consisted of a 2-min period at 95°C followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min. The program ended with one cycle at 72°C for 5 min. The products were purified using a Minielute Gel extraction kit (Qiagen, USA) according to the manufacturer’s directions. The primer sets used for HA and NA were designed by Hoffmann et al. [18]. The genes of internal proteins were amplified using the primer sets designed by Liu et al. [19].
Sequencing and phylogenetic analysis
The purified PCR products were then sequenced using the primers described above and a CEQ dye terminal cycle sequence quick start kit (Beckman Coulter, USA) and a CEQ 2000 automatic sequencer (Beckman Coulter, USA). Multiple sequence alignments were made with Clustal W (http://clustalw.ddbj.nig.ac.jp/top-j.html). The phylogenetic trees were generated with the Phylip program by the neighbor-joining method. The robustness of the groupings was tested by bootstrap resampling. The neighbor-joining trees with 1000 replicates were used in the bootstrap. The phylogenetic trees were drawn using TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). In addition to the strains described above, the nucleotide sequence data for influenza virus genes in the DNA Data Bank were used for phylogenetic analysis.
Results
Six influenza viruses were isolated from 1,413 fecal samples of terns that were collected on Heron Island, Australia, in December 2004. All were identified as subtype H2N5 by HI and NI tests. These isolates were designated A/tern/Australia/1/04 (H2N5), A/tern/Australia/91/04 (H2N5), A/tern/Australia/1363/04 (H2N5), A/tern/Australia/1378/04 (H2N5), and A/tern/Australia/1402/04 (H2N5). All eight segments of these viruses were partially sequenced. Since there are no data on the N5 NA gene sequence belonging to the Eurasian lineage in the DNA Data Bank, NA segments of 11 Eurasian isolates of the N5 subtype in our laboratory were also sequenced. The sequence data have been deposited in the DNA Data Bank (AB266379–AB266386, AB275403–AB275415, AB275614-AB275632, AB275858-AB275865, AB270594–AB270600, and AB270746–AB270749).
The alignment of sequences showed that six isolates were extremely homologous (no less than 99% homology in all segments), indicating that these terns were infected with the same strain. Upon analysis of A/tern/Australia/1/04 (H2N5) [Tn/Aus/1/04 (H2N5)] with the alignment search tool (BLAST) available from the DNA Data Bank, it was found that the HA, NA, PB2, PB1, PA, and NS gene segments of Tn/Aus/1/04 (H2N5) were most closely related to the genes of A/mallard duck/Netherlands/13/99 (H2N2) (97%), A/duck/Mongolia/149/03 (H10N5) (90%), A/pheasant/Ireland/PV18/97 (H9N2) (97%), A/duck/Shanghai/38/01 (H5N1) (96%), A/chicken/Guandong/174/04 (H5N1) (96%), and A/duck/Nanchang/8-174/00 (H3N6) (96%), respectively. The NP and M gene segments of Tn/Aus/1/04 (H2N5) were most homologous with the NP and M of A/guillemot/Sweden/3/00 (H6N2) (99% and 97% identity, respectively). The nucleotide sequence analysis, thus, revealed that they originated from different sources, except for the NP and M genes (Table 1).
By phylogenetic analysis, the M gene of the present isolates was found to belong to the American lineage of avian influenza viruses and the other genes to the Eurasian lineage (Fig. 1). The NA genes were of Eurasian avian lineage together with those of 11 strains isolated in Asia. All the isolates from Oceania including the present isolates and the isolates of the 1970s and 1980s belonged either to the American or to the Eurasian lineage. The PA genes belonged to a sublineage of highly pathogenic H5N1 avian influenza viruses isolated in 2004 in Asia including Japan.
Phylogenetic trees for the eight gene segments of influenza viruses. Nucleotides 477–1021 (545 bp) of HA, 539–1381 (843 bp) of NA, 1508–2161 (654 bp) of PB2, 1168–1734 (654 bp) of PB1, 764–1202 (438 bp) of PA, 1135–1433 (299 bp) of NP, 60–929 (870 bp) of M, and 38–528 (491 bp) of NS were used for the phylogenetic analysis. The phylogenetic trees were constructed by the neighbor-joining method and bootstrap testing (n = 1000). The phylogenetic trees of HA and NA were rooted to A/tern/South Africa/61 (H5N3) and A/duck/Hong Kong/27408/79 (H5N3), respectively. The phylogenetic trees of the other genes were rooted to B/Lee. Bar, 0.1 substitution per site. Australian strains sequenced in the present study are highlighted in gray. Australian viruses in the 1970s and 1980s are underlined. ALB, Alberta; DE, Delaware; MT, Massachusetts; NJ, New Jersey; NY, New York; NZL, New Zealand
Discussion
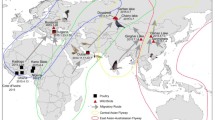
The virological surveillance of avian influenza in feral birds has become important, since H5N1 HPAI viruses have been repeatedly isolated [12, 20]. To obtain information on the influenza virus in feral water birds in the southern hemisphere, we conducted virological surveillance by isolating avian influenza viruses from fecal samples of terns in Australia. Phylogenetic analysis of the genes of the present isolates revealed the M gene segment to be of American origin and the other gene segments to be of Eurasian origin (Fig. 1). The genes, with the exception of the NP and M genes, have different origins (Table 1). These results suggest the present isolates to be generated by multiple reassortments, similar to strain A/guillemot/Sweden/3/00 (H6N2) reported by Wallensten et al. [21]. Migratory birds have variable migration routes [7]. Some of the North–South routes the birds fly are the East Asia-Australian flyway and the American flyway, and these routes overlap around Alaska. In Alaska, migratory birds congregate during the breeding season in summer. Therefore, the reassortment of Eurasian and American lineages might arise during nesting in Alaska. The NP and M genes of the present isolates showed high sequence identity with those of A/guillemot/Sweden/3/00 (H6N2) [21]. Wallensten et al. [21] found that the NP gene of A/guillemot/Sweden/3/00 (H6N2) is Eurasian and the M gene is American. PB2 and HA genes showed high sequence identity with genes of strains from Ireland and the Netherlands, respectively. Sweden, Ireland, and the Netherlands are on the East Atlantic flyway. The eastern and western ends of the East Atlantic flyway overlap with the western end of the East Asia-Australian flyway and the eastern end of the American flyway, respectively. Migratory birds, thus, come in contact with those taking different flyways. In these migratory flyways, the present isolates might have been provided with genes of European lineage. Many wader species of the Northern Hemisphere are long-distance intercontinental migrants [7], and the flyways provide overlapping areas. No genes of the present isolates from Australia are related to the genes of isolates in Australia and New Zealand in the 1970s and 1980s (Fig. 1 and Table 1). All the Oceanian isolates analyzed in the present study belonged to the American or Eurasian lineage. These results indicate that Oceanian isolates including the present isolates do not perpetuate in feral water birds in Australia but carry viruses from the Northern hemisphere.
The PA genes of the present isolates belonged to the lineage of H5N1 HPAI isolates in Asia including Japan in 2004, indicating a common origin. Previous surveillance by our laboratory indicated influenza viruses perpetuated in ducks nesting in Siberia to have provided genes for the emergence of the H5N1 virus in Hong Kong in 1997 [15]. On the basis of the present results, it is suggested that the PA gene of common origin was provided to HPAI H5N1 viruses isolated in Asia such as A/chicken/Guandong/174/04 (H5N1) and the present isolates by migratory birds. It is assumed that migratory birds might provide the PA gene of avirulent viruses to HPAI H5N1 strains and the present isolates. However, as there have been cases of the reintroduction of H5N1 HPAI viruses from domestic to wild birds, possibility of the introduction of the PA gene from H5N1 virus strain into the present tern virus cannot be excluded. Although it is difficult to identify how the present isolates were generated, it is presumed that while the viruses of different origin were mixed somewhere on the migratory routes, the present isolates were generated as a result of reassortment among various strains. Migratory birds fly to Australia through Asia, the site of outbreaks of H5N1 influenza, and the present isolates in Australia and H5N1 viruses isolated in Asia have a common PA gene, and H5N1 HPAI outbreaks have occurred in Indonesia located close to Australia [8]. Therefore, it is impossible to exclude the possibility that migratory birds will carry H5N1 viruses to Australia in the future although no HPAI virus was isolated from the fecal samples of terns in this study.
The isolation of H2 subtype influenza viruses from wild birds in Australia has not been reported. The H2 subtype influenza virus caused an influenza pandemic in humans 40 years ago. As young people are immunologically naive of this subtype, the possible reemergence of the H2 influenza virus causing a pandemic in the near future cannot be ruled out. Therefore, extensive surveillance studies of free-flying birds are needed to provide information to prepare for future pandemics.
References
V.S. Hinshaw, G.M. Air, A.J. Gibbs, L. Graves, B. Prescott, D. Karunakaran, J. Virol. 42, 865–872 (1982)
Y. Kawaoka, S. Yamnikova, T.M. Chambers, D.K. Lvov, R.G. Webster, Virology 179, 759–767 (1990). doi:https://doi.org/10.1016/0042–6822(90)90143-F
C. Rohm, N. Zhou, J. Suss, J. Mackenzie, R.G. Webster, Virology 217, 508–516 (1996). doi:https://doi.org/10.1006/viro.1996.0145
R.A. Fouchier, V. Munster, A. Wallensten, T.M. Bestebroer, S. Herfst, D. Smith, G.F. Rimmelzwaan, B. Olsen, A.D. Osterhaus, J. Virol. 79, 2814–2822 (2005). doi:https://doi.org/10.1128/JVI.79.5.2814-2822.2005
G.B. Abenes, K. Okazaki, H. Fukushi, H. Kida, E. Honda, K. Yagyu, M. Tsuji, H. Sato, E. Ono, R. Yanagawa, N. Yamauchi, Nippon Juigaku Zasshi 44, 703–708 (1982)
R.G. Webster, W.J. Bean, O.T. Gorman, T.M. Chambers, Y. Kawaoka, Microbiol. Rev. 56, 152–179 (1992)
B. Olsen, V.J. Munster, A. Wallensten, J. Waldenstrom, A.D. Osterhaus, R.A. Fouchier, Science 312, 384–388 (2006). doi:https://doi.org/10.1126/science.1122438
K.S. Li, Y. Guan, J. Wang, G.J. Smith, K.M. Xu, L. Duan, A.P. Rahardjo, P. Puthavathana, C. Buranathai, T.D. Nguyen, A.T. Estoepangestie, A. Chaisingh, P. Auewarakul, H.T. Long, N.T. Hanh, R.J. Webby, L.L. Poon, H. Chen, K.F. Shortridge, K.Y. Yuen, R.G. Webster, J.S. Peiris, Nature 430, 209–213 (2004). doi:https://doi.org/10.1038/nature02746
M. Mase, K. Tsukamoto, T. Imada, K. Imai, N. Tanimura, K. Nakamura, Y. Yamamoto, T. Hitomi, T. Kira, T. Nakai, M. Kiso, T. Horimoto, Y. Kawaoka, S. Yamaguchi, Virology 332, 167–176 (2005). doi:https://doi.org/10.1016/j.virol.2004.11.016
M.F. Ducatez, C.M. Olinger, A.A. Owoade, Z. Tarnagda, M.C. Tahita, A. Sow, S. De Landtsheer, W. Ammerlaan, J.B. Ouedraogo, A.D. Osterhaus, R.A. Fouchier, C.P. Muller, J. Gen. Virol. 88, 2297–2306 (2007). doi:https://doi.org/10.1099/vir.0.82939-0
N. Kishida, Y. Sakoda, N. Isoda, K. Matsuda, M. Eto, Y. Sunaga, T. Umemura, H. Kida, Arch. Virol. 150, 1383–1392 (2005). doi:https://doi.org/10.1007/s00705-004-0473-x
J. Liu, H. Xiao, F. Lei, Q. Zhu, K. Qin, X.W. Zhang, X.L. Zhang, D. Zhao, G. Wang, Y. Feng, J. Ma, W. Liu, J. Wang, G.F. Gao, Science 309, 1206 (2005). doi:https://doi.org/10.1126/science.1115273
K.M. Sturm-Ramirez, T. Ellis, B. Bousfield, L. Bissett, K. Dyrting, J.E. Rehg, L. Poon, Y. Guan, M. Peiris, R.G. Webster, J. Virol. 78, 4892–4901 (2004). doi:https://doi.org/10.1128/JVI.78.9.4892-4901.2004
T. Ito, K. Okazaki, Y. Kawaoka, A. Takada, R.G. Webster, H. Kida, Arch. Virol. 140, 1163–1172 (1995). doi:https://doi.org/10.1007/BF01322743
K. Okazaki, A. Takada, T. Ito, M. Imai, H. Takakuwa, M. Hatta, H. Ozaki, T. Tanizaki, T. Nagano, A. Ninomiya, V.A. Demenev, M.M. Tyaptirganov, T.D. Karatayeva, S.S. Yamnikova, D.K. Lvov, H. Kida, Arch. Virol. 145, 885–893 (2000). doi:https://doi.org/10.1007/s007050050681
Y. Sakoda, T. Ito, K. Okazaki, A. Takada, Y. Ito, K. Tamai, M. Okamatsu, K. Shortridge, R. Webster, H. Kida, Proceedings of the International Conference on Options for the Control of Influenza V 1263, 674–677 (2004)
H. Kida, R. Yanagawa, Zentralbl. Bakteriol. [Orig. A] 244, 135–143 (1979)
E. Hoffmann, J. Stech, Y. Guan, R.G. Webster, D.R. Perez, Arch. Virol. 146, 2275–2289 (2001). doi:https://doi.org/10.1007/s007050170002
J. Liu, K. Okazaki, H. Ozaki, Y. Sakoda, Q. Wu, F. Chen, H. Kida, Avian Pathol. 32, 551–560 (2003). doi:https://doi.org/10.1080/0307-9450310001596728
K.M. Sturm-Ramirez, D.J. Hulse-Post, E.A. Govorkova, J. Humberd, P. Seiler, P. Puthavathana, C. Buranathai, T.D. Nguyen, A. Chaisingh, H.T. Long, T.S. Naipospos, H. Chen, T.M. Ellis, Y. Guan, J.S. Peiris, R.G. Webster, J. Virol. 79, 11269–11279 (2005). doi:https://doi.org/10.1128/JVI.79.17.11269–11279.2005
A. Wallensten, V.J. Munster, J. Elmberg, A.D. Osterhaus, R.A. Fouchier, B. Olsen, Arch. Virol. 150, 1685–1692 (2005). doi:https://doi.org/10.1007/s00705-005-0543-8
Acknowledgments
We appreciate the considerable support that Dr. Graem Laver’s family has provided in the collection of fecal samples in Australia. The present study was supported by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kishida, N., Sakoda, Y., Shiromoto, M. et al. H2N5 influenza virus isolates from terns in Australia: genetic reassortants between those of the Eurasian and American lineages. Virus Genes 37, 16–21 (2008). https://doi.org/10.1007/s11262-008-0235-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-008-0235-z