Abstract
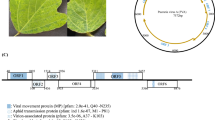
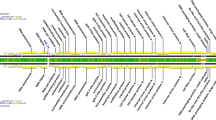
The complete DNA sequence of the mildly virulent Gallid herpesvirus type 2 strain CU-2 was determined and consists of 176,922 bp with an overall gene organization typical of class E herpesviruses. Phylogenetically, this strain partitions in its own branch between the virulent strains RB-1B, Md11, and Md5, and the vaccine strain CVI988. Overall, the genome of CU-2 is more similar to that of CVI988, with identically sized unique short regions of 11,651 bp. As in CVI988, an insertion of 177 bp was identified in the overlapping genes encoding the Meq, RLORF6, and 23 kDa proteins within the repeat long region of the genome. A total of 15 single nucleotide polymorphisms (SNPs) common to both CU-2 and CVI988, and not occurring in virulent strains, were identified in the genes encoding UL29, UL45, UL50, UL52, LORF10, RLORF14a, RLORF12, Meq(RLORF7), 23kDa, ICP4, US3, and two hypothetical proteins MDV071.4 and MDV076.4. Each gene encoding UL29 and Meq contained two SNPs. Only one major open reading frame (ORF) encoding UL41, the virus host shutoff (VHS) ribonuclease, was disrupted in the CU-2 genome. An additional cytosine after the 25 codon is predicted to produce a truncated protein of 97 aa. Since GaHV-2 mutants lacking UL41 have been reported to retain their virulence, other factors are likely responsible for the low virulence of CU-2. It is largely suspected that SNPs in common with CVI988 along with the insertions in the Meq loci are responsible for its phenotype. Conversely, we identified 43 nonsynonymous mutations (within 23 genes) that may contribute to the virulence of CU-2. These SNPs are shared exclusively with all sequenced virulent strains (Md5, Md11, and RB-1B) and not present within the CVI988 genome. Although most occur in proteins of unknown function, a significant percentage is in proteins involved in virion assembly.




Similar content being viewed by others
References
R. Borenshtein, I. Davidson, Development of the hot spot-combined PCR assay for detection of retroviral insertions into Marek’s disease virus. J. Virol. Methods 82, 119–127 (1999)
P. Brunovskis, L.F. Velicer, The Marek’s disease virus (MDV) unique short region: alphaherpesvirus-homologous, fowlpox virus-homologous, and MDV-specific genes. Virology 206, 324–338 (1995)
B.W. Calnek, J. Fabricant, K.A. Schat, K.K. Murthy, Pathogenicity of low-virulence Marek’s disease viruses in normal versus immunologically compromised chickens. Avian Dis. 21, 346–358 (1977)
K.S. Chang, K. Ohashi, M. Onuma, Diversity (polymorphism) of the Meq gene in the attenuated Marek’s disease virus (MDV) serotype 1 and MDV-transformed cell lines. J. Vet. Med. Sci. 64, 1097–1101 (2002)
K.S. Chang, K. Ohashi, M. Onuma, Suppression of transcription activity of the MEQ protein of oncogenic Marek’s disease virus serotype 1 (MDV1) by L-MEQ of non-oncogenic MDV1. J. Vet. Med. Sci. 64, 1091–1095 (2002)
A.E. Churchill, L.N. Payne, R.C. Chubb, Marek’s disease immunization against Marek’s disease using a live attenuated virus. Lancet 1, 610–611 (1969)
F. Corpet, Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988)
I. Davidson, R. Borenshtain, In vivo events of retroviral long terminal repeat integration into Marek’s disease virus in commercial poultry: detection of chimeric molecules as a marker. Avian Dis. 45, 102–121 (2001)
H.J. Delecluse, W. Hammerschmidt, Status of Marek’s disease virus in established lymphoma cell lines: herpesvirus integration is common. J. Virol. 67, 82–92 (1993)
H.J. Delecluse, S. Schuller, W. Hammerschmidt, Latent Marek’s disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 12, 3277–3286 (1993)
H. Deng, S. Dewhurst, Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J. Virol. 72, 320–329 (1998)
R.C. Edgar, MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004)
D. Endoh, M. Ito, K.O. Cho, Y. Kon, T. Morimura, M. Hayashi, M. Kuwabara, Retroviral sequence located in border region of short unique region and short terminal repeat of Md5 strain of Marek’s disease virus type 1. J. Vet. Med. Sci. 60, 227–235 (1998)
J. Fabricant, M. Ianconescu, B.W. Calnek, Comparative effects of host and viral factors on early pathogenesis of Marek’s disease. Infect. Immun. 16, 136–144 (1977)
M. Hohl, I. Dunand-Sauthier, L. Staresincic, P. Jaquier-Gubler, F. Thorel, M. Modesti, S.G. Clarkson, O.D. Scharer, Domain swapping between FEN-1 and XPG defines regions in XPG that mediate nucleotide excision repair activity and substrate specificity. Nucleic Acids Res. 35, 3053–3063 (2007)
R.J. Isfort, D. Robinson, H.J. Kung, Purification of genomic sized herpesvirus DNA using pulse-field electrophoresis. J. Virol. Methods 27, 311–317 (1990)
R.J. Isfort, Z. Qian, D. Jones, R.F. Silva, R. Witter, H.J. Kung, Integration of multiple chicken retroviruses into multiple chicken herpesviruses: herpesviral gD as a common target of integration. Virology 203, 125–133 (1994)
D. Jones, R. Isfort, R. Witter, R. Kost, H.J. Kung, Retroviral insertions into a herpesvirus are clustered at the junctions of the short repeat and short unique sequences. Proc. Natl. Acad. Sci. USA 90, 3855–3859 (1993)
D. Jones, P. Brunovskis, R. Witter, H.J. Kung, Retroviral insertional activation in a herpesvirus: transcriptional activation of US genes by an integrated long terminal repeat in a Marek’s disease virus clone. J. Virol. 70, 2460–2467 (1996)
K. Karbstein, J.A. Doudna, GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J. Mol. Biol. 356, 432–443 (2006)
C. Kaschka-Dierich, K. Nazerian, R. Thomssen, Intracellular state of Marek’s disease virus DNA in two tumour-derived chicken cell lines. J. Gen. Virol. 44, 271–280 (1979)
K. Katoh, K. Kuma, H. Toh, T. Miyata, MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005)
R. Kost, D. Jones, R. Isfort, R. Witter, H.J. Kung, Retrovirus insertion into herpesvirus: characterization of a Marek’s disease virus harboring a solo LTR. Virology 192, 161–169 (1993)
S. Kumar, K. Tamura, M. Nei, MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 5, 150–163 (2004)
L.F. Lee, E.D. Kieff, S.L. Bachenheimer, B. Roizman, P.G. Spear, B.R. Burmester, K. Nazerian, Size and composition of Marek’s disease virus deoxyribonucleic acid. J. Virol. 7, 289–294 (1971)
L.F. Lee, P. Wu, D. Sui, D. Ren, J. Kamil, H.J. Kung, R.L. Witter, The complete unique long sequence and the overall genomic organization of the GA strain of Marek’s disease virus. Proc. Natl. Acad. Sci. USA 97, 6091–6096 (2000)
S.I. Lee, M. Takagi, K. Ohashi, C. Sugimoto, M. Onuma, Difference in the Meq gene between oncogenic and attenuated strains of Marek’s disease virus serotype 1. J. Vet. Med. Sci. 62, 287–292 (2000)
M. Margulies, M. Egholm, W.E. Altman, S. Attiya, J.S. Bader, L.A. Bemben, J. Berka, M.S. Braverman, Y.J. Chen, Z. Chen, S.B. Dewell, L. Du, J.M. Fierro, X.V. Gomes, B.C. Godwin, W. He, S. Helgesen, C.H. Ho, G.P. Irzyk, S.C. Jando, M.L. Alenquer, T.P. Jarvie, K.B. Jirage, J.B. Kim, J.R. Knight, J.R. Lanza, J.H. Leamon, S.M. Lefkowitz, M. Lei, J. Li, K.L. Lohman, H. Lu, V.B. Makhijani, K.E. McDade, M.P. McKenna, E.W. Myers, E. Nickerson, J.R. Nobile, R. Plant, B.P. Puc, M.T. Ronan, G.T. Roth, G.J. Sarkis, J.F. Simons, J.W. Simpson, M. Srinivasan, K.R. Tartaro, A. Tomasz, K.A. Vogt, G.A. Volkmer, S.H. Wang, Y. Wang, M.P. Weiner, P. Yu, R.F. Begley, J.M. Rothberg, Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005)
M. Niikura, J. Dodgson, H. Cheng, Direct evidence of host genome acquisition by the alphaherpesvirus Marek’s disease virus. Arch. Virol. 151, 537–549 (2005)
W. Okazaki, H.G. Purchase, B.R. Burmester, Protection against Marek’s disease by vaccination with a herpesvirus of turkeys. Avian Dis. 14, 413–429 (1970)
K. Osterrieder, J.-F. Vautherot, The genome content of Marek’s disease-like viruses, in Marek’s Disease: An Emerging Problem, ed. by F. Davison, V. Nair (Elsevier, Oxford, UK, 2004)
J.D. Retief, Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 132, 243–258 (2000)
H.-J. Rhiza, B. Bauer, Circular forms of viral DNA in Marek’s disease virus-transformed lymphoblastoid cells. Arch. Virol. 72, 211–216 (1982)
B.H. Rispens, H. van Vloten, N. Mastenbroek, H.J. Maas, K.A. Schat, Control of Marek’s disease in the Netherlands. I. Isolation of an avirulent Marek’s disease virus (strain CVI 988) and its use in laboratory vaccination trials. Avian Dis. 16, 108–125 (1972)
B. Roizman, in The Herpesviruses, ed. by B. Roizman (Plenum, London, 1982), p. 1–23
B. Roizman, in Fields Virology, 4th edn., ed. by B.N. Fields, D.M. Knipe, P.M. Howley (Lippincott-Raven Press, New York, 1996), p. 2221–2230
B. Roizman, A.E. Sears, in Fields Virology, ed. by B.N. Fields, D.M. Knipe, P.M. Howley (Lippincott-Raven Press, New York, 1996), p. 2231–2295
N. Ross, M.M. Binns, M. Sanderson, K.A. Schat, Alterations in DNA sequence and RNA transcription of the BamHI-H fragment accompany attenuation of oncogenic Marek’s disease herpesvirus. Virus Genes 7, 33–51 (1993)
A.A. Schaffer, L. Aravind, T.L. Madden, S. Shavirin, J.L. Spouge, Y.I. Wolf, E.V. Koonin, S.F. Altschul, Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29, 2994–3005 (2001)
K.A. Schat, B.W. Calnek, Characterizations of an apparently non-oncogenic Marek’s disease virus. J. Natl. Cancer Inst. 60, 1141–1146 (1978)
C.E. Shamblin, N. Greene, V. Arumugaswami, R.L. Dienglewicz, M.S. Parcells, Comparative analysis of Marek’s disease virus (MDV) glycoprotein-, lytic antigen pp38- and transformation antigen Meq-encoding genes: association of Meq mutations with MDVs of high virulence. Vet. Microbiol. 102, 147–167 (2004)
R.F. Silva, I. Gimeno, Oncogenic Marek’s disease viruses lacking the 132 base pair repeats can still be attenuated by serial in vitro cell culture passages. Virus Genes 34, 87–90 (2007)
R.F. Silva, R.L. Witter, Genomic expansion of Marek’s disease virus DNA is associated with serial in vitro passage. J. Virol. 54, 690–696 (1985)
R.F. Silva, S.M. Reddy, B. Lupiani, Expansion of a unique region in the Marek’s disease virus genome occurs concomitantly with attenuation but is not sufficient to cause attenuation. J. Virol. 78, 733–740 (2004)
M.W. Smith, B.W. Calnek, Effect of virus pathogenicity on antibody production in Marek’s disease. Avian Dis. 17, 727–736 (1973)
M.W. Smith, B. Calnek, Comparative features of low-virulence and high-virulence Marek’s disease infections. Avian Pathol. 3, 229–246 (1974)
S.J. Spatz, R.F. Silva, Polymorphisms in the repeat long regions of oncogenic and attenuated pathotypes of Marek’s disease virus 1. Virus Genes 35, 41–53 (2007)
S.J. Spatz, R.F. Silva, Sequence determination of variable regions within the genomes of gallid herpesvirus-2 pathotypes. Arch. Virol. 152, 1665–1678 (2007)
S.J. Spatz, L. Petherbridge, Y. Zhao, V. Nair, Comparative full-length sequence analysis of oncogenic and vaccine (Rispens) strains of Marek’s disease virus. J. Gen. Virol. 88, 1080–1096 (2007)
S.J. Spatz, Y. Zhao, L. Petherbridge, L.P. Smith, S.J. Baigent, V. Nair, Comparative sequence analysis of a highly oncogenic but horizontal spread-defective clone of Marek’s disease virus. Virus Genes 35, 753–766 (2007)
B. Taddeo, B. Roizman, The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J. Virol. 80, 9341–9345 (2006)
A. Tanaka, S. Silver, M. Nonoyama, Biochemical evidence of the nonintegrated status of Marek’s disease virus DNA in virus-transformed lymphoblastoid cells of chicken. Virology 88, 19–24 (1978)
E.R. Tulman, C.L. Afonso, Z. Lu, L. Zsak, D.L. Rock, G.F. Kutish, The genome of a very virulent Marek’s disease virus. J. Virol. 74, 7980–7988 (2000)
R.L. Witter, Characteristics of Marek’s disease viruses isolated from vaccinated commercial chicken flocks: association of viral pathotype with lymphoma frequency. Avian Dis. 27, 113–132 (1983)
R.L. Witter, Increased virulence of Marek’s disease virus field isolates. Avian Dis. 41, 149–63 (1997)
R.L. Witter, B.W. Calnek, C. Buscaglia, I.M. Gimeno, K.A. Schat, Classification of Marek’s disease viruses according to pathotype: philosophy and methodology. Avian Pathol. 34, 75–90 (2005)
Acknowledgments
We thank Jeremy Volkening for his contributions in all stages of this research. Jeremy has outstanding skills in bioinformatics, scientific writing, and computer graphics. This research was funded by the United States Department of Agriculture CRIS program (project number 6612-32000-043).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Spatz, S.J., Rue, C.A. Sequence determination of a mildly virulent strain (CU-2) of Gallid herpesvirus type 2 using 454 pyrosequencing. Virus Genes 36, 479–489 (2008). https://doi.org/10.1007/s11262-008-0213-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-008-0213-5