Abstract
Probably due to its phytogeographic position, the grasslands in the North Adriatic Karst are among the richest grasslands in the world and harbour the highest small-scale density of plant species found in terrestrial habitats. Different moisture and soil conditions determine distinct vegetation types, such as meadows, composed by mesophyllous plants and on mesic conditions, and semi-natural pastures, composed by sclerophyllous plants and on oligotrophic conditions. Even though plants in these two vegetation types differ in some of their attributes, their functional and phylogenetic relationships remain to be tested. We used a dataset comprising 180 species, in which 48 plots in the meadows and 52 plots in the pastures had been sampled, and tested the phylogenetic and functional relationships between the two vegetation types. The pastures contained more original species, but both the pastures and the meadows are expected to be important to increase biodiversity at landscape level. Different community assembly processes occurred in the two vegetation types, with limiting similarity leading to functional overdispersion in the meadows and environmental filtering leading to functional clustering in the pastures. Overall, traits were convergent, leading to a clustered phylogenetic structure in the meadows, probably due to pair-wise competition, and an overdispersed phylogenetic structure in the pastures, where species from different clades were filtered by the oligotrophic conditions. Transhumance may have contributed to the random pattern of trait diversity we found across the nodes of the phylogenetic tree.

Similar content being viewed by others
References
Ackerly D (2009) Conservatism and diversification of plant functional traits: evolutionary rates versus phylogenetic signal. Proc Natl Acad Sci USA 106:19699–19706
Andrič M, Willis KJ (2003) The phytogeographical regions of Slovenia: a consequence of natural environmental variation or prehistoric human activity? J Ecol 91:807–821
ARSO (2009) Meteorološki letopis—mesečna višina padavin. http://www.arso.gov.si/vreme/podnebje/meteorološki%20letopis/2009pad_vis.pdf. Accessed 6 Dec 2012
Beaulieu JM, Ree RH, Cavender-Bares J, Weiblen GD, Donoghue MJ (2012) Synthesizing phylogenetic knowledge for ecological research. Ecology 93:S4–S13
Bell CD, Soltis DE, Soltis PS (2010) The age and diversification of the angiosperms re-revisited. Am J Bot 97:1296–1303
Birks HJB, Line JM, Persson T (1990) Quantitative estimation of human impact on cultural landscape development. In: Birks HH, Kaland PE, Moe D (eds) The cultural landscape—past, present and future. Cambridge University Press, Cambridge, pp 229–240
Blomberg SP, Garland T Jr, Ives A (2003) Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57:717–745
Braun-Blanquet J (1964) Pflanzensoziologie: grundzüge der vegetationskunde. Springer, Wien
Carvalho GH, Cianciaruso MV, Batalha MA (2010) Plantminer: a web tool for checking and gathering plant species taxonomic information. Environ Model Softw 25:815–816
Clarke KR, Warwick RM (1998) A taxonomic distinctness index and its statistical properties. J Appl Ecol 35:523–531
Clarke KR, Warwick RM (2001) A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar Ecol Prog Ser 216:265–278
Collins PM, Davis BAS, Kaplan JO (2012) The mid-Holocene vegetation of the Mediterranean region and southern Europe, and comparison with the present day. J Biogeogr 38:1848–1861
Cornelissen JHC, Lavorel S, Gernier E, Diaz S, Buchmann N, Gurvich DE, Reich PB, ter Steege H, Morgan HD, van der Heijden MGA, Pausas JG, Poorter H (2003) A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust J Bot 51:335–380
Cousins SAO, Eriksson A (2001) Plant species occurrences in a rural hemiboreal landscape: effects of remnant habitats, site history, topography and soil. Ecography 24:461–469
de Beaulieu JL, Miras Y, Andrieu-Ponel V, Guiter F (2005) Vegetation dynamics in north-western Mediterranean regions: instability of the Mediterranean bioclimate. Plant Biosyst 139:114–126
Diacon-Bolli J, Dalang T, Holderegger R, Bürgi M (2012) Heterogeneity fosters biodiversity: linking history and ecology of dry calcareous grasslands. Basic Appl Ecol 13:641–653
Dray S, Dufour AB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10
Harvey PH, Pagel M (1991) The comparative method in evolutionary biology. Oxford University Press, Oxford
Jombart T, Dray S (2008) Adephylo: exploratory analyses for the phylogenetic comparative method. http://cran.r-project.org/web/packages/adephylo/index.html. Accessed 6 Dec 2012
Jombart T, Pavoine S, Devillard S, Pontier D (2010) Putting phylogeny into the analysis of biological traits: a methodological approach. J Theor Biol 264:693–701
Kaligarič M (1997) Rastlinstvo Primorskega krasa in Slovenske Istre-Travniki in pašniki.-Zgodovinsko društvo za južno Primorsko. ZRS-RS Koper, Koper
Kaligarič M, Poldini L (1996) New contribution on the typology of the vegetation of dry grasslands Scorzoneretalia villosae H-ić 1975 in the North Adriatic Karst. Gortania 19:119–148
Kaligarič M, Culiberg M, Kramberger B (2006) Recent vegetation history of the North Adriatic grasslands: expansion and decay of an anthropogenic habitat. Folia Geobot 41:241–258
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464
Kleyer M, Bekker RM, Knevel IC, Bakker JP, Thompson K, Sonnenschein M, Poschlod P, van Groenendael JM, Klimes L, Klimesová J, Klotz S, Rusch GM, Hermy M, Adriaens D, Boedeltje G, Bossuyt B, Endels P, Götzenberger L, Hodgson JG, Jackel AK, Dannemann A, Kühn I, Kunzmann D, Ozinga WA, Römermann C, Stadler M, Schlegelmilch J, Steendam HJ, Tackenberg O, Wilmann B, Cornelissen JHC, Eriksson O, Garnier E, Fitter A, Peco B (2008) The LEDA Traitbase: a database of plant life-history traits of Northwest European flora. J Ecol 96:1266–1274
Klotz S, Kühn I, Durka W (2002) BIOLFLOR: eine Datenbank mit biologisch-ökologischen Merkmalen zur flora von Deutschland. Schriftenr Vegetationsk 38:1–334
Köppen W (1931) Grundriss der Klimakunde. Gruyter, Berlin
Martinčič A, Wraber T, Jogan N, Podobnik A, Turk B, Vreš B (2007) Mala flora Slovenije: kjuč za določanje praprotnic in semenk. Tehniška založba Slovenije, Ljubljana
Nee S, May RM (1997) Extinction and the loss of evolutionary history. Science 278:692–694
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2012) Vegan: community ecology package. http://CRAN.R-project.org/package=vegan. Accessed 6 Dec 2012
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Pasquale G, Martino P, Mazzoleni S (2004) Forest history in the Mediterranean region. In: Mazzoleni S, Pasquale G, Mulligan M, Martino P, Rego F (eds) Recent dynamics of the Mediterranean vegetation and landscape. Wiley, West Sussex, pp 13–20
Pausas JG, Verdú M (2010) The jungle of methods for evaluating phenotypic and phylogenetic structure of communities. Bioscience 60:614–625
Pavoine S, Ollier S, Dufour AB (2005) Is the originality of a species measurable? Ecol Lett 8:579–586
Pavoine S, Vallet J, Dufour AB, Gachet S, Daniel H (2009) On the challenge of treating various types of variables: application for improving the measurement of functional diversity. Oikos 118:391–402
Pavoine S, Baguette M, Bonsall MB (2010) Decomposition of trait diversity among the nodes of a phylogenetic tree. Ecol Monogr 80:485–507
Petchey OL, Gaston KJ (2002) Functional diversity (FD), species richness and community composition. Ecol Lett 5:402–411
Pipenbaher N, Kaligarič M, Škornik S (2011) Floristic and functional comparison of Karst pastures and Karst meadows from the North Adriatic Karst. Acta Carsologica 40:515–525
Poldini L (1989) La vegetazione del Carso isontino e triestino. Lint, Trieste
Quézel P, Medail F (2003) Ecologie et biogeography des forests du basin Mediterranean. Elsevier, Paris
Ruhfel BR, Bittrich V, Bove CP, Gustafsson MHG, Philbrick CT, Rutishauser R, Xi Z, Davis CC (2011) Phylogeny of the Clusioid clade (Malpighiales): evidence from the plastid and mitochondrial genomes. Am J Bot 98:306–325
Šilc U (2010) Synanthropic vegetation: pattern of various disturbances on life history traits. Acta Bot Croat 69:215–227
Škornik S, Vidrih M, Kaligarič M (2010) The effect of grazing pressure on species richness, composition and productivity in North Adriatic Karst pastures. Plant Biosyst 144:55–364
Srivastava DS, Cadotte MC, MacDonald AAM, Marushia RG, Mirotchnick N (2012) Phylogenetic diversity and the functioning of ecosystems. Ecol Lett 15:637–648
R Development Core Team (2012) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna. URL: http://www.r-project.org
Watts D (2004) Quaternary biotic interactions in Slovenia and adjacent regions: the vegetation. In: Griffiths HIB (ed) Balkan biodiversity: pattern and process in the European hotspots. Kluwer, Dordrecht, pp 69–78
Webb CO (2000) Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am Nat 156:145–155
Webb CO, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology. Ann Rev Ecol Syst 33:475–505
Wurdack KJ, Davis CC (2009) Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot 96:1551–1570
Acknowledgments
We are grateful to ‘Fundação de Amparo à Pesquisa do Estado de São Paulo’, for financial support, and to Mitja Kaligarič, for support and helpful comments on the paper.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Appendix 1
See Table 5
References
Ajani Y, Ajani A, Cordes JM, Watson MF, Downie SR (2008) Phylogenetic analysis of nrDNA ITS sequences reveals relationships within five groups of Iranian Apiaceae subfamily Apioideae. Taxon 57: 383–401.
Barres L, Vilatersana R, Molero J, Susanna A, Galbany-Casals M (2011) Molecular phylogeny of Euphorbia subg. Esula sect. Aphyllis (Euphorbiaceae) inferred from nrDNA and cpDNA markers with biogeographic insights. Taxon 60: 705–720.
Bell CD, Soltis DE, Soltis PS (2010) The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97: 1296–1303.
Bennett JR, Matthews S (2006) Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am. J. Bot. 93: 1039–1051.
Bouchenak-Khelladi Y, Salamin N, Savolainen V, Forest F, Bank M, Chase MW, Hodkinson TR (2008) Large multi-gene phylogenetic trees of the grasses (Poaceae): progress towards complete tribal and generic level sampling. Mol. Phylogenet. Evol. 47: 488–505.
Cai YF, Li SW, Chen M, Jiang MF, Liu Y, Xie YF, Sun Q, Jiang HZ, Yin NW, Wang L, Zhang R, Huang CL, Lei K (2010) Molecular phylogeny of Ranunculaceae based on rbc L sequences. Biologia 65: 997–1003.
Calviño CI, Downie SR (2007) Circumscription and phylogeny of Apiaceae subfamily Saniculoideae based on chloroplast DNA sequences. Mol. Phylogenet. Evol 44: 175–191.
Catalán P, Kellogg EA, Olmstead RG (1997) Phylogeny of Poaceae subfamily Pooideae based on chloroplast ndhF gene sequences. Mol. Phylogenet. Evol 8: 150–166.
Chase MW, Reveal JL, Fay MF (2009) A subfamilial classification for the expanded asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae. Bot. J. Linn. Soc. 161: 132–136.
Dobeš C, Paule J (2010) A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): Implications for its geographic origin, phylogeography and generic circumscription. Mol. Phylogenet. Evol 56: 156–175.
Doyle JJ, Chappill JA, Bailey DC, Kajita T (2000) Towards a comprehensive phylogeny of legumes: evidence from rbcL sequences and non-molecular data. In: Herendeen PS and Bruneau A. Adv. Legum. Systemat. 9: 1–20.
Downie SR, Katz-Downie DS, Spalik K (2000a) A phylogeny of Apiaceae tribe Scandiceae: evidence from nuclear ribosomal DNA internal transcribed spacer sequences. Am. J. Bot. 87: 76–95.
Downie SR, Katz-Downie DS, Watson MF (2000b) A phylogeny of the flowering plant family Apiaceae based on chloroplast DNA rpl16 and rpoC1 intron sequences: towards a suprageneric classification of subfamily Apioideae. Am. J. Bot. 87: 273–292.
Downie SR, Ramanath S, Katz-Downie DS, Llanas E (1998) Molecular systematics of Apiaceae subfamily Apioideae: phylogenetic analyses of nuclear ribosomal DNA internal transcribed spacer and plastid RPO C1 intron sequences. Am. J. Bot. 85: 563–591.
Drew BT, Sytsma KJ (2012) Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am. J. Bot. 99: 933–953.
Eddie WMM, Shulkina T, Gaskin J, Haberle RC, Jansen RK (2003) Phylogeny of Campanulaceae s. str. inferred from ITS sequences of nuclear ribosomal DNA. Ann. Mo. Bot. Gard. 90: 554–575.
Ellison NW, Liston A, Steiner JJ, Williams WM, Taylor NL (2006) Molecular phylogenetics of the clover genus (Trifolium – Leguminosae). Mol. Phylogenet. Evol 39: 688–705.
Emadzade K, Gehrke B, Linder HP, Hörandl E (2011) The biogeographical history of the cosmopolitan genus Ranunculus L. (Ranunculaceae) in the temperate to meridional zones. Mol. Phylogenet. Evol 58: 4–21.
Eriksson T, Donoghue MJ, Hibbs MS (1998) Phylogenetic analysis of Potentilla using DNA sequences of nuclear ribosomal internal transcribed spacers (ITS), and implications for the classification of Rosoideae (Rosaceae). Plant Syst. Evol. 211: 155–179.
Font M, Garnatje T, Garcia-Jacas N, Susanna A (2002) Delineation and phylogeny of Centaurea sect. Acrocentron based on DNA sequences: a restoration of the genus Crocodylium and indirect evidence of introgression. Plant Syst. Evol. 234: 15–26.
Garcia-Jacas N, Susanna A, Garnatje T, Vilatersana R (2001) Generic delimitation and phylogeny of the subtribe Centaureinae (Asteraceae): a combined nuclear and chloroplast DNA analysis. Ann. Bot. 87: 503–515.
Garcia-Jacas N, Uysal T, Romashchenko K, Suárez-Santiago VN, Ertuğrul K, Susanna A (2006) Centaurea revisited: a molecular survey of the Jacea group. Ann. Bot. 98: 741–753.
Goertzen LR, Cannone JJ, Gutell RR, Jansen RK (2003) ITS secondary structure derived from comparative analysis: implications for sequence alignment and phylogeny of the Asteraceae. Mol. Phylogenet. Evol 29: 216–234.
Hendrichs M, Oberwinkler F, Begerow D, Bauer R (2004) Carex, subgenus Carex (Cyperaceae) – A phylogenetic approach using ITS sequences. Plant Syst. Evol. 246: 89–107.
Hörandl E, Paun O, Johansson JT, Lehnebach C, Armstrong T, Chen L, Lockhart P (2005) Phylogenetic relationships and evolutionary traits in Ranunculus s.l. (Ranunculaceae) inferred from ITS sequence analysis. Mol. Phylogenet. Evol 36: 305–327.
Horn JW, Ee BW, Morawetz JJ, Riina R, Steinmann VW, Berry PE, Wurdack KJ (2012) Phylogenetics and the evolution of major structural characters in the giant genus Euphorbia L. (Euphorbiaceae). Mol. Phylogenet. Evol 63: 305–326.
Hungerer KB, Kadereit JW (1998) The phylogeny and biogeography of Gentiana L. sect. Ciminalis (Adans.) Dumort.: A historical interpretation of distribution ranges in the European high mountains. Perspect. Plant Ecol. 1: 121–135.
Inda LA, Pimentel M, Chase MW (2012) Phylogenetics of tribe Orchideae (Orchidaceae: Orchidoideae) based on combined DNA matrices: inferences regarding timing of diversification and evolution of pollination syndromes. Ann. Bot. 110: 71–90.
Käss E, Wink M (1997) Phylogenetic relationships in the Papilionoideae (family Leguminosae) based on nucleotide sequences of cpDNA (rbcL) and ncDNA (ITS 1 and 2). Mol. Phylogenet. Evol 8: 65–88.
Kilian N, Gemeinholzer B, Lack HW (2009) Cichorieae. In: Funk VA, Susanna A, Stuessy TF, and Bayer RJ. Systematics, evolution, and biogeography of Compositae. International Association for Plant Taxonomy, Vienna, pp. 343–383.
Kim DK, Kim JS, Kim JH (2012) The phylogenetic relationships of Asparagales in Korea based on five plastid DNA regions. J. Plant Biol. 55: 325–341.
Li QQ, Zhou SD, He XJ, Yu Y, Zhang YC, Wei XQ (2010) Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann. Bot. 106: 709–733.
Liang H (1997) The phylogenetic reconstruction of the grass family (Poaceae) using matK gene sequences. PhD Thesis, Virginia Polytechnic Institute and State University, Blacksburg.
Lindqvist C, Albert VA (2002) Origin of the Hawaiian endemic mints within North American Stachys (Lamiaceae). Am. J. Bot. 89: 1709–1724.
Manzanilla V, Bruneau A (2012) Phylogeny reconstruction in the Caesalpinieae grade (Leguminosae) based on duplicated copies of the sucrose synthase gene and plastid markers. Mol. Phylogenet. Evol 65: 149–162.
Mathews S, Tsai RC, Kellogg EA (2000) Phylogenetic structure in the grass family (Poaceae): evidence from the nuclear gene phytochrome B. Am. J. Bot. 87: 96–107.
McDill J, Repplinger M, Simpson BB, Kadereit JW (2009) The phylogeny of Linum and Linaceae subfamily Linoideae, with implications for their systematics, biogeography, and evolution of heterostyly. Syst. Bot. 34: 386–405.
Meszaros S, Laet J, Smets E (1996) Phylogeny of temperate Gentianaceae: a morphological approach. Syst. Bot. 21: 153–168.
Moon HK, Smets E, Huysmans S (2010) Phylogeny of tribe Mentheae (Lamiaceae): The story of molecules and micromorphological characters. Taxon 59: 1065–1076.
Morrone O, Aagesen L, Scataglini MA, Salariato DL, Denham SS, Chemisquy MA, Sede SM, Giussani LM, Kellogg EA, Zuloaga FO (2012) Phylogeny of the Paniceae (Poaceae: Panicoideae): integrating plastid DNA sequences and morphology into a new classification. Cladistics 28: 333–356.
Panero JL, Funk VA (2008) The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol. Phylogenet. Evol 47: 757–782.
Pardo C, Cubas P, Tahiri H (2004) Molecular phylogeny and systematics of Genista (Leguminosae) and related genera based on nucleotide sequences of nrDNA (ITS region) and cpDNA (trnL-trnF intergenic spacer). Plant Syst. Evol. 244: 93–119.
Paun O, Lehnebach C, Johansson JT, Lockhart P, Hörandl E (2005) Phylogenetic relationships and biogeography of Ranunculus and allied genera (Ranunculaceae) in the Mediterranean region and in the European Alpine system. Taxon 54: 911–930.
Potter D, Eriksson T, Evans RC, Oh S, Smedmark JEE, Morgan DR, Kerr M, Robertson KR, Arsenault M, Dickinson TA, Campbell CS (2007) Phylogeny and classification of Rosaceae. Plant Syst. Evol. 266: 5–43.
Quintanar A, Castroviejo S, Catalán P (2007) Phylogeny of the tribe Aveneae (Pooideae, Poaceae) inferred from plastid trnT-F and nuclear ITS sequences. Am. J. Bot. 94: 1554–1569.
Rahn K (1996) A phylogenetic study of the Plantaginaceae. Bot. J. Linn. Soc. 120: 145–198.
Rønsted N, Chase MW, Albach DC, Bello MA (2002) Phylogenetic relationships within Plantago (Plantaginaceae): evidence from nuclear ribosomal ITS and plastid trnL-F sequence data. Bot. J. Linn. Soc. 139: 323–338.
Ruhfel BR, Bittrich V, Bove CP, Gustafsson MHG, Philbrick CT, Rutishauser R, Xi Z, Davis CC (2011) Phylogeny of the Clusioid clade (Malpighiales): evidence from the plastid and mitochodrial genomes. Am. J. Bot. 98: 306–325.
Ryding O (2010) Pericarp structure and phylogeny of tribe Mentheae (Lamiaceae). Plant Syst. Evol. 285: 165–175.
Samuel R, Gutermann W, Stuessy TF, Ruas CF, Lack HW, Termetsberger K, Talavera S, Hermanowski B, Ehrendorfer F (2006) Molecular phylogenetics reveals Leontodon (Asteraceae, Lactuceae) to be diphyletic. Am. J. Bot. 93: 1193–1205.
Soza VL (2010) Diversification of Galium within tribe Rubieae (Rubiaceae): evolution of breeding systems, species complexes, and gene duplication. PhD Thesis, University of Washington, Washington.
Soza VL, Olmstead RG (2010) Molecular systematics of tribe Rubieae (Rubiaceae): evolution of major clades, development of leaf-like whorls, and biogeography. Taxon 59: 755–771.
Steele KP, Ickert-Bond SM, Zarre S, Wojciechowski MF (2010) Phylogeny and character evolution in Medicago (Leguminosae): evidence from analyses of plastid trnK/matK and nuclear GA3ox1 sequences. Am. J. Bot. 97: 1142–1155.
Susanna A, Garcia-Jacas N, Hidalgo O, Vilatersana R, Garnatje T (2006) The Cardueae (Compositae) revisited: insights from ITS, trnL-trnF, and matK nuclear and chloroplast DNA analysis. Ann. Mo. Bot. Gard. 93: 150–171.
Taskova RM, Gotfredsen CH, Jensen SR (2006) Chemotaxonomy of Veroniceae and its allies in the Plantaginaceae. Phytochemistry 67: 286–301.
Teerawatananon A, Jacobs SWL, Hodkinson TR (2011) Phylogenetics of Panicoideae (Poaceae) based on chloroplast and nuclear DNA sequences. Telopea 13: 115–142.
Těšitel J, Říha P, Svobodová Š, Malinová T, Štech M (2010) Phylogeny, life history evolution and biogeography of the Rhinanthoid Orobanchaceae. Folia Geobotanica 45: 347–367.
Walker JB, Sytsma KJ, Treutlein J, Wink M (2004) Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am. J. Bot. 91: 1115–1125.
Watson LE, Evans TM, Boluarte T (2000) Molecular phylogeny and biogeography of tribe Anthemideae (Asteraceae), based on Chloroplast Gene ndhF. Mol. Phylogenet. Evol 15: 59–69.
Wink M (2003) Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64: 3–19.
Wink M, Kaufmann M (1996) Phylogenetic relationships between some members of the subfamily Lamioideae (family Labiatae) inferred from nucleotide sequences of the rbcL gene. Bot. Acta 109: 139–148.
Wojciechowski MF (2003) Reconstructing the phylogeny of legumes (Leguminosae): an early 21st century perspective. In Klitgaard BB and Bruneau A. Adv. Legum. Systemat. 10: 5–35.
Wojciechowski MF, Sanderson MJ, Steele KP, Liston A (2000) Molecular phylogeny of the “Temperate Herbaceous Tribes” of Papilionoid legumes: a supertree approach. In Herendeen PS and Bruneau A. Adv. Legum. Systemat. 9: 277–298.
Wu ZQ, Ge S (2012) The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol. Phylogenet. Evol 62: 573–578.
Wurdack KJ, Davis CC (2009) Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am. J. Bot. 96: 1551–1570.
Zhang W (2000) Phylogeny of the grass family (Poaceae) from rpl16 Intron sequence data. Mol. Phylogenet. Evol 15: 135–146.
Zimmermann NFA, Ritz CM, Hellwig FH (2010) Further support for the phylogenetic relationships within Euphorbia L. (Euphorbiaceae) from nrITS and trnL–trnF IGS sequence data. Plant Syst. Evol. 286: 39–58.
Appendix 2
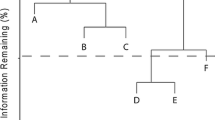
See Fig. 2
Rights and permissions
About this article
Cite this article
Pipenbaher, N., Škornik, S., de Carvalho, G.H. et al. Phylogenetic and functional relationships in pastures and meadows from the North Adriatic Karst. Plant Ecol 214, 501–519 (2013). https://doi.org/10.1007/s11258-013-0185-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11258-013-0185-y