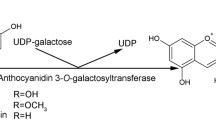
Abstract
Anthocyanins are responsible for the red color in red-fleshed kiwifruits. In most of these cyanidin 3-O-xyl-galactoside is the predominant anthocyanin. This requires a second glycosylation modification which attaches the UDP-xylose to the cyanidin 3-O-galactoside. We report here the functional characterization of two second glycosyltransferases from ‘Hongyang’ kiwifruit, AcUFGT6b and AcUFGT7c. These were found to cluster with other plant GGTs. In vitro, recombinant AcUFGT6b recognized 3-O-glycosylated anthocyanins and UDP-xylose, while AcUFGT7c showed no or negligible activity. In tobacco leaves, co-expression of AcMYBF110, AcUFGT3a and AcUFGT6b resulted in the accumulation of three new products: cyanidin 3-O-xyl-galactoside, cyanidin 3-O-xyl-glucoside and cyanidin 3-O-xyl-rutoside. RNAi assays showed that a lack of AcUFGT6b and AcUFGT7c in immature fruits of Actinidia arguta prevents the formation of cyanidin 3-O-xyl-galactoside. Also, the content of cyanidin 3-O-xyl-galactoside was obviously increased when AcUFGT6b was injected together with AcMYBF110 and AcUFGT6b7c-RNAi. In contrast, for AcUFGT7c, as well as in vitro, we show no activity in either tobacco leaves or kiwifruit. Both AcUFGT6b and AcUFGT7c are located in the ER and their promoters can be positively activated by AcMYBF110, indicating the functional differences of them are due neither to their cell localization nor to the upstream regulation of AcMYBF110. Interestingly, the deletion in the C-terminal of AcUFGT7c is likely to change its 3D protein structure compared with AcUFGT6b protein, resulting in it having no capacity to attract the UDP sugar moiety. These results suggest that AcUFGT6b, but not AcUFGT7c, is responsible for the end-product of the anthocyanins biosynthesis pathway in red-fleshed kiwifruit.
Key message
AcUFGT6b attracts the UDP-xylose to the cyanidin 3-O-galactoside generating cyanidin 3-O-xyl-galactoside, and positively regulated by the AcMYBF110.






Similar content being viewed by others
References
Ahmed SS, Gong ZH, Ji JJ, Yin YX, Xiao HJ, Khan MA, Rehman A, Ahmad I (2012) Construction of the intermediate vector pVBG2307 by incorporating vital elements of expression vectors pBI121 and pBI221. Genet Mol Res 11(3):3091–3104
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13(3):99–102
Andersen OM, Jordheim M (2006) The anthocyanins. In: Andersen O, Markham KR (eds) Flavonoids. CRC Press, Boca Raton, pp 471–551
Bi X, Zhang J, Chen C, Zhang D, Li P, Ma F (2014) Anthocyanin contributes more to hydrogen peroxide scavenging than other phenolics in apple peel. Food Chem 152:205–209
Bieza K, Lois R (2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126(3):1105–1115
Breton C, Fournel-Gigleux S, Palcic MM (2012) Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol 22(5):540–549
Brugliera F, Holton TA, Stevenson TW, Farcy E, Lu CY, Cornish EC (1994) Isolation and characterization of a cDNA clone corresponding to the Rt locus of Petunia hybrida. Plant J 5(1):81–92
Cheng J, Wei G, Zhou H, Gu C, Vimolmangkang S, Liao L, Han Y (2014) Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol 166(2):1044–1058
Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13
Hiromoto T, Honjo E, Tamada T, Noda N, Kazuma K, Suzuki M, Kuroki R (2013) Crystal structure of UDP-glucose:anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. J Synchrotron Radiat 20(Pt 6):894–898
Hiromoto T, Honjo E, Noda N, Tamada T, Kazuma K, Suzuki M, Blaber M, Kuroki R (2015) Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. Protein Sci 24(3):395–407
Hugueney P, Provenzano S, Verries C, Ferrandino A, Meudec E, Batelli G, Merdinoglu D, Cheynier V, Schubert A, Ageorges A (2009) A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol 150(4):2057–2070
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18(9):477–483
Jiang Y, Liu C, Yan D, Wen X, Liu Y, Wang H, Dai J, Zhang Y, Liu Y, Zhou B, Ren X (2017) MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. J Exp Bot 68(5):1055–1069
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Kobayashi S, Ishimaru M, Ding CK, Yakushiji H, Goto N (2001) Comparison of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci 160(3):543–550
Lee Y, Yoon HR, Paik YS, Liu JR, Chung WI, Choi G (2005) Reciprocal regulation of Arabidopsis UGT78D2 and BANYULS is critical for regulation of the metabolic flux of anthocyanidins to condensed tannins in developing seed coats. J Plant Biol 48(4):356–370
Li JQ, Li XW, Soejarto DD (2007) Actinidiaceae (Flora of China). Acta Phytotax 45:633–660
Li W, Liu Y, Zeng S, Xiao G, Wang G, Wang Y, Peng M, Huang H (2015) Gene expression profiling of development and anthocyanin accumulation in kiwifruit (Actinidia chinensis) based on transcriptome sequencing. PLoS ONE 10(8):e0136439
Li B, Xia Y, Wang Y, Qin G, Tian S (2017) Characterization of genes encoding key enzymes involved in anthocyanin metabolism of kiwifruit during storage period. Front Plant Sci 8:341
Liu Y, Zhou B, Qi Y, Chen X, Liu C, Liu Z, Ren X (2017) Expression differences of pigment structural genes and transcription factors explain flesh coloration in three contrasting kiwifruit cultivars. Front Plant Sci 8:1507
Liu Y, Zhou B, Qi Y, Liu C, Liu Z, Ren X (2018) Biochemical and functional characterization of AcUFGT3a, a galactosyltransferase involved in anthocyanin biosynthesis in the red-fleshed kiwifruit (Actinidia chinensis). Physiol Plant 162(4):409–426
Liu Y, Qi Y, Chen X, He H, Liu Z, Zhang Z, Ren Y, Ren X (2019) Phenolic compounds and antioxidant activity in red- and in green-fleshed kiwifruits. Food Res Int 116:291–301
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25(4):402–408
Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Belanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T, Lancet D, Louisot P, Magdalou J, Chowdhury JR, Ritter JK, Schachter H, Tephly TR, Tipton KF, Nebert DW (1997) The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7(4):255–269
Man YP, Wang YC, Li ZZ, Jiang ZW, Yang HL, Gong JJ, He SS, Wu SQ, Yang ZQ, Zheng J, Wang ZY (2015) High-temperature inhibition of biosynthesis and transportation of anthocyanins results in the poor red coloration in red-fleshed Actinidia chinensis. Physiol Plant 153(4):565–583
Montefiori M, Comeskey DJ, Wohlers M, McGhie TK (2009) Characterization and quantification of anthocyanins in red kiwifruit (Actinidia spp.). J Agric Food Chem 57(15):6856–6861
Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC (2011) Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J 65(1):106–118
Morita Y, Hoshino A, Kikuchi Y, Okuhara H, Ono E, Tanaka Y, Fukui Y, Saito N, Nitasaka E, Noguchi H, Iida S (2005) Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J 42(3):353–363
Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51(6):1126–1136
Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25(6):1396–1405
Paquette SM, Jensen K, Bak S (2009) A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases (http://www.P450.kvl.dk). Phytochemistry 70 (17–18):1940–1947
Petroni K, Pilu R, Tonelli C (2014) Anthocyanins in corn: a wealth of genes for human health. Planta 240(5):901–911
Rose A, Glassgen WE, Hopp W, Seitz HU (1996) Purification and characterization of glycosyltransferases involved in anthocyanin biosynthesis in cell-suspension cultures of Daucus carota L. Planta 198(3):397–403
Santos-Buelga C, Mateus N, De Freitas V (2014) Anthocyanins. Plant pigments and beyond. J Agric Food Chem 62(29):6879–6884
Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T (2005) UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J Biol Chem 280(2):899–906
Sun W, Liang L, Meng X, Li Y, Gao F, Liu X, Wang S, Gao X, Wang L (2016) Biochemical and molecular characterization of a flavonoid 3-O-glycosyltransferase responsible for anthocyanins and flavonols biosynthesis in freesia hybrida. Front Plant Sci 7:410
Sun JJ, Wang YC, Chen XS, Gong XJ, Wang N, Ma L, Qiu YF, Wang YL, Feng SQ (2017) Effects of methyl jasmonate and abscisic acid on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Organ Culture 130:227–237
Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142(3):1216–1232
Veeriah S, Kautenburger T, Habermann N, Sauer J, Dietrich H, Will F, Pool-Zobel BL (2006) Apple flavonoids inhibit growth of HT29 human colon cancer cells and modulate expression of genes involved in the biotransformation of xenobiotics. Mol Carcinog 45(3):164–174
Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T (2013) The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol 162(2):885–896
Wang N, Zhang ZY, Jiang SH, Xu HF, Wang YC, Feng SQ, Chen XS (2016) Synergistic effects of light and temperature on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Organ Cult 127:217–227
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126(2):485–493
Yang HL, Wang YC, Jiang ZW, Huang HW (2009) Construction of cDNA library of ‘Hongyang’ kiwifruit and analysis of F3H expression. HEREDITAS 31(12):1265–1272
Yang J, Jiang ZW, Wang YC (2010) Cloning and expression of dihydroflavonol 4-reductase in Actinidia chinensis var. rufopulpa. J Wuhan Bot Res 28(6):673–681
Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, Wangwattana B, Yamazaki M, Saito K (2012) Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J 69(1):154–167
Yu Y, Xu W, Wang J, Wang L, Yao W, Yang Y, Xu Y, Ma F, Du Y, Wang Y (2013) The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol 200(3):834–846
Zhang L, Man YP, Jiang ZW, Wang YC (2012) Cloning and expression of anthocyanin pathway genes, AcCHS and AcLDOX, in Actinidia chinensis. Acta Hortic Sin 39(11):2124–2132
Zhao J, Dixon RA (2010) The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci 15(2):72–80
Zhao D, Tao J (2015) Recent advances on the development and regulation of flower color in ornamental plants. Front Plant Sci 6:261
Zhou H, Lin-Wang K, Wang H, Gu C, Dare AP, Espley RV, He H, Allan AC, Han Y (2015) Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J 82(1):105–121
Acknowledgements
This work was supported by the National Seeds Innovation Engineering: Kiwifruit Breeding Innovation (Grant No: C000082). The authors thank professor Wan Jianmin for kindly providing the ER-located marker labeled with mCherry. The authors also thank help of Qianqian Shi for the experiments.
Author information
Authors and Affiliations
Contributions
YL designed the experiments, performed the research, wrote and revised this manuscript; JL, YQ and AZ performed the subcellular localization assay and helped revised manuscript. ZL designed the experiments and provided all samples tested; XR designed the experiments, discussed results, and revised this manuscript. All authors have participated in this research and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Ali R. Alan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11240_2019_1622_MOESM1_ESM.xlsx
Supplementary material 1 (XLSX 29 kb) Table S1 Primers used in this study. Table S2 Substrate specificity of AcUFGT6b and AcUFGT7c.
11240_2019_1622_MOESM2_ESM.tif
Supplementary material 2 (TIFF 6974 kb) Figure S1 The predicted 3D structures of AcUFGT6b (a) and AcUFGT7c (b) and their corresponding distribution of α-helix and β-sheet (c). The magenta boxed region indicates the PSPG motif.
Rights and permissions
About this article
Cite this article
Liu, Y., Liu, J., Qi, Y. et al. Identification and characterization of AcUFGT6b, a xylosyltransferase involved in anthocyanin modification in red-fleshed kiwifruit (Actinidia chinensis). Plant Cell Tiss Organ Cult 138, 257–271 (2019). https://doi.org/10.1007/s11240-019-01622-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-019-01622-6