Abstract
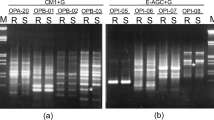
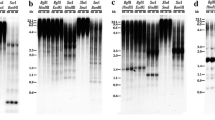
In order to develop more specific markers that characterize particular regions of the pea genome, the data on nucleotide sequences of RAPD fragments were used for choosing more extended primers, which may be helpful in amplifying a fragment corresponding to the particular DNA region. Of the 14 STS markers obtained from 14 polymorphic RAPD fragments, 12 were polymorphic, i.e., they are SCAR markers that can be used in genetic analysis. The transition from complex RAPD spectra to amplification of a particular SCAR marker substantially facilitates analysis of large samples for the presence or absence of the examined fragment. Inheritance of the developed SCAR markers was studied in F1 and F2. SCAR markers were used to identify various pea lines, cultivars, and mutants. It was established that the study of amplification of STS markers in various pea genotypes at varying temperatures of annealing and the comparison with amplification of the original RAPD fragments in the same genotypes provide an approach for analysis of RAPD polymorphism origin.
Similar content being viewed by others
REFERENCES
Mohan, M., Nair, S., Bhagwat, A., et al., Genome Mapping, Molecular Markers and Marker-Assisted Selection in Crop Plants, Mol. Breed., 1997, vol. 3, pp. 87–103.
Williams, J.G.K., Kubelik, A.R., Livak, K.J., et al., DNA Polymorphisms Amplified by Arbitrary Primers Are Useful As Genetic Markers, Nucleic Acids Res., 1990, vol. 18, pp. 6531–6535.
Paran, J. and Michelmore, R.W., Development of Reliable PCR-Based Markers Linked to Downy Mildew Resistance Genes in Lettuce, Theor. Appl. Genet., 1993, vol. 85, pp. 985–993.
Deng, Z., Huang, S., Xiao, S.Y., and Gmitter, F.G., Jr. Development and Characterization of SCAR Markers Linked to the Citrus Tristeza Virus Resistance Gene from Poncirus trifoliata, Genome, 1997, vol. 40, pp. 697–704.
Koveza, O.V., Kokaeva, Z.G., Konovalov, F.A., and Gostimskii, S.A., Identification and Mapping of Polymorphic RAPD Markers of Pea (Pisum sativum L.) Genome, Rus. J. Genet., 2005, vol. 41, no.3, pp. 262–268.
Rychlik, W. and Rhoads, R.E., A Computer Program for Choosing Optimal Oligonucleotides for Filter Hybridization, Sequencing and In Vitro Amplification of DNA, Nucleic Acids Res., 1989, vol. 17, pp. 8543–8551.
Weng, C., Kubisiak, T.L., and Stine, M., SCAR Markers in a Longleaf Pine × Slash Pine F1 Family, Forest Genet., 1998, vol. 5, no.4, pp. 239–247.
Weber, J., Diez, J., Selosse, M.A., et al., SCAR Markers to Detect Mycorrhizas of an American Laccaria bicolor Strain Inoculated in European Douglas-Fir Plantations, Mycorrhiza, 2002, vol. 12, no.1, pp. 19–27.
Newton, C.R., Graham, A., Heptinstall, L.E., et al., Analysis of Any Point Mutation in DNA: The Amplification Refractory Mutation System (ARMS), Nucleic Acids Res., 1989, vol. 17, no.7, pp. 2503–2516.
Zykova, E.S., Patrushev, L.I., Kayushin, A.L., et al., New Allele-Specific Primers for Detecting the Leiden Mutation in Exon 10 of the Factor V Gene in Thrombophilia, Bioorg. Khim., 1997, vol. 23, pp. 205–210.
Patrushev, L.I., Zykova, E.S., Kayushin, A.L., et al., New Test System for DNA Diagnosis and Identification of Homozygous and Heterozygous Point Mutations, Bioorg. Khim., 1998, vol. 24, pp. 194–200.
Kwok, S., Kellog, D.E., McKinney, N., et al., Effects of Primer-Template Mismatches on the Polymerase Chain Reaction: Human Immunodeficiency Virus Type 1 Model Studies, Nucleic Acids Res., 1990, vol. 18, pp. 999–1005.
Hernandez, P., Dorado, G., Cabrera, A., et al., Rapid Verification of Wheat-Hordeum Introgressions by Direct Staining of SCAR, STS, and SSR Amplicons, Genome, 2002, vol. 45, pp. 198–203.
Author information
Authors and Affiliations
Additional information
__________
Translated from Genetika, Vol. 41, No. 11, 2005, pp. 1522–1530.
Original Russian Text Copyright © 2005 by Koveza, Gostimsky.
Rights and permissions
About this article
Cite this article
Koveza, O.V., Gostimsky, S.A. Development and Study of SCAR Markers in Pea (Pisum sativum L.). Russ J Genet 41, 1254–1261 (2005). https://doi.org/10.1007/s11177-005-0226-2
Received:
Issue Date:
DOI: https://doi.org/10.1007/s11177-005-0226-2