Abstract
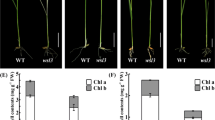
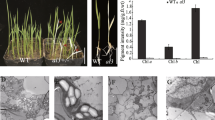
In higher plant chloroplasts, the plastid-encoded RNA polymerase (PEP) consists of four catalytic subunits and numerous nuclear-encoded accessory proteins, including pTAC10, an S1-domain-containing protein. In this study, pTAC10 knockout lines were characterized. Two ptac10 mutants had an albino phenotype and severely impaired chloroplast development. The pTAC10 genomic sequence fused to a four-tandem MYC tag driven by its own promoter functionally complemented the ptac10-1 mutant phenotype. pTAC10 was present in both the chloroplast stroma and thylakoids. Two-dimensional blue native polyacrylamide gel electrophoresis (BN-PAGE), and immunoblotting assays showed that pTAC10:MYC co-migrates with one of the PEP core subunits, RpoB. A comprehensive investigation of the plastid gene expression profiles by quantitative RT-PCR revealed that, compared with wild-type plants, the abundance of PEP-dependent plastid transcripts is severely decreased in the ptac10-1 mutant, while the amount of plastid transcripts exclusively transcribed by NEP either barely changes or even increases. RNA blot analysis confirmed that PEP-dependent chloroplast transcripts, including psaB, psbA and rbcL, substantially decrease in the ptac10-1 mutant. Immunoblotting showed reduced accumulation of most chloroplast proteins in the ptac10 mutants. These data indicate the essential role of pTAC10 in plastid gene expression and plastid development. pTAC10 interacts with chloroplast-targeted casein kinase 2 (cpCK2) in vitro and in vivo and can be phosphorylated by Arabidopsis cpCK2 in vitro at sites Ser95, Ser396 and Ser434. RNA-EMSA assays showed that pTAC10 is able to bind to the psbA, atpE and accD transcripts, suggesting a non-specific RNA-binding activity of pTAC10. The RNA affinity of pTAC10 was enhanced by phosphorylation and decreased by the amino acid substitution Ser434-Ala of pTAC10. These data show that pTAC10 is essential for plastid gene expression in Arabidopsis and that it can be phosphorylated by cpCK2.









Similar content being viewed by others
References
Allison LA, Simon LD, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15:2802–2809
Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustun S, Melzer M, Petersen K, Lein W, Bornke F (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22:1498–1515
Baginsky S, Gruissem W (2009) The chloroplast kinase network: new insights from large-scale phosphoproteome profiling. Mol Plant 2:1141–1153
Baginsky S, Tiller K, Link G (1997) Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba). Plant Mol Biol 34:181–189
Baginsky S, Tiller K, Pfannschmidt T, Link G (1999) PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Mol Biol 39:1013–1023
Berry JO, Yerramsetty P, Zielinski AM, Mure CM (2013) Photosynthetic gene expression in higher plants. Photosynth Res 117(1–3):91–120
Chang SH, Lee S, Um TY, Kim JK, Choi YD, Jang G (2017) pTAC10, a key subunit of plastid-encoded RNA polymerase, promotes chloroplast development. Plant Physiol. https://doi.org/10.1104/pp.17.00248
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
De Santis-MacIossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rüdiger W, Koop HU, Herrmann RG (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J 18:477–489
Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S (2006) Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol 142:993–1003
Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiolgy 157:1733–1745
Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H (2008) An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 53:924–934
Ghavidel A, Schultz MC (2001) TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106:575–584
Gilkerson J, Perez-Ruiz JM, Chory J, Callis J (2012) The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biol 12:102
Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16:4041–4048
Hess WR, Prombona A, Fieder B, Subramanian AR, Bo¨rner T (1993) Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning nonchloroplast- encoded RNA polymerase. EMBO J 12:563–571
Hu J, Bogorad L (1990) Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci USA 87:1531–1535
Ingelsson B, Vener AV (2012) Phosphoproteomics of Arabidopsis chloroplasts reveals involvement of the STN7 kinase in phosphorylation of nucleoid protein pTAC16. FEBS Lett 586(9):1265–1271
Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T (2005) A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J 42:133–144
Jeon Y, Jung HJ, Kang H, Park YI, Lee SH, Pai HS (2012) S1 domain-containing STF modulates plastid transcription and chloroplast biogenesis in Nicotiana benthamiana. New Phytol 193:349–363
Krause K, Maier RM, Kofer W, Krupinska K, Herrmann RG (2000) Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol Genet Genomics 263:1022–1030
Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31:171–188
Link G (1996) Green life: control of chloroplast gene transcription. Bioessays 18:465–471
Lisitsky I, Liveanu V, Schuster G (1995) RNA-binding characteristics of a ribonucleoprotein from spinach chloroplast. Plant Physiol 107:933–941
Litchfield DW (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J 369:1–15
Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49:557–577
Loza-Tavera H, Vargas-Suárez M, Díaz-Mireles E, Torres-Márquez ME, González de la Vara LE, Moreno-Sánchez R, Gruissem W (2006) Phosphorylation of the spinach chloroplast 24 kDa RNA-binding protein (24RNP) increases its binding to petD and psbA 3′ untranslated regions. Biochimie 88:1217–1228
Lu Q, Ding S, Reiland S, Rödiger A, Roschitzki B, Xue P, Gruissem W, Lu C, Baginsky S (2015) Identification and characterization of chloroplast casein kinase II from Oryza sativa (rice). J Exp Bot 66(1):175–187
Melonek J, Matros A, Trösch M, Mock HP, Krupinska K (2012) The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell 24:3060–3073
Mulekar JJ, Huq E (2013) Expanding roles of protein kinase CK2 in regulating plant growth and development. J Exp Bot 65:2883–2893
Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K (2008) A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20:3148–3162
Niefind K, Guerra B, Ermakowa I, Issinger OG (2001) Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J 20:5320–5331
Ogrzewalla K, Piotrowski M, Reinbothe S, Link G (2002) The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur J Biochem 269:3329–3337
Pfalz J, Pfannschmidt T (2013) Essential nucleoid proteins in early chloroplast development. Trends Plant Sci 18:186–194
Pfalz J, Liere K, Kandlbinder A, DietzK J, Oelmuller R (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18:176–197
Pfalz J, Holtzegel U, Barkan A, Weisheit W, Mittag M, Pfannschmidt T (2015) ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol 206:1024–1037
Pfannschmidt T, Link G (1994) Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol 25:69–81
Pfannschmidt T, Ogrzewalla K, Baginsky S, Sickmann A, Meyer HE, Link G (2000) The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.): integration of a prokaryotic core into a larger complex with organelle-specific functions. Eur J Biochem 267:253–261
Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150:889–903
Salinas P, Fuentes D, Vidal E, Jordana X, Echeverria M, Holuigue L. 2006. An extensive survey of CK2 alpha and beta subunits in Arabidopsis: multiple isoforms exhibit differential subcellular localization. Plant Cell Physiol 47:1295–1308
Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 6:283–290
Schönberg A, Bergner E, Helm S, Agne B, Dünschede B, Schünemann D, Schutkowski M, Baginsky S (2014) The peptide microarray “ChloroPhos1.0” identifies new phosphorylation targets of plastid casein kinase II (pCKII) in Arabidopsis thaliana. PLoS ONE 9:e108344
Schröter Y, Steiner S, Matthäi K, Pfannschmidt T (2010) Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics 10:2191–2204
Schweer J, Türkeri H, Link B, Link G (2010) AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. Plant J 62:192–202
Silhavy D, Maliga P (1998) Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr Genet 33:340–344
Steiner S, Schroter Y, Pfalz J, Pfannschmidt T (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157:1043–1055
Stöckel J, Bennewitz S, Hein P, Oelmüller R (2006) The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol 141:870–878
Suzuki JY, Ytterberg AJ, Beardslee TA, Allison LA, Wijk KJ, Maliga P (2004) Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J 40:164–172
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vargas-Suárez M, Castro-Sánchez A, Toledo-Ortiz G, González de la Vara LE, García E, Loza-Tavera H (2013) Protein phosphorylation regulates in vitro spinach chloroplast petD mRNA 3′-untranslated region stability, processing, and degradation. Biochimie 95:400–409
Wakasugi T, Tsudzuki T, Sugiura M (2001) The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynth Res 70(1):107–118
Williams-Carrier R, Zoschke R, Belcher S, Pfalz J, Barkan A (2014) A major role for the plastid-encoded RNA polymerase complex in the expression of plastid transfer RNAs. Plant Physiol 164:239–248
Yagi Y, Ishizaki Y, Nakahira Y, Tozawa Y, Shiina T (2012) Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc Natl Acad Sci USA 109:7541–7546
Yang Z, Shang Z, Wang L, Lu Q, Wen X, Chi W, Zhang L, Lu C (2015) Purine biosynthetic enzyme ATase2 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Photosynth Res 126(2–3):285–300
Yu QB, Jiang Y, Chong K, Yang ZN (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59:1011–1023
Yu QB, Lu Y, Ma Q, Zhao TT, Huang C, Zhao HF, Zhang XL, Lv RH, Yang ZN (2013) TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiol Plant 148:408–421
Yu QB, Huang C, Yang ZN (2014) Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front Plant Sci 5:316
Zhang HD, Cui YL, Huang C, Yin QQ, Qin XM, Xu T, He XF, Zhang Y, Li ZR, Yang ZN (2015) PPR protein PDM1/SEL1 is involved in RNA editing and splicing of plastid genes in Arabidopsis thaliana. Photosynth Res 126(2–3):311–321
Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, Peng L, Zhang L, Lu C (2013) Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25:2925–2943
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgements
We thank ABRC Bioresources, which kindly provided the transgenic Arabidopsis lines (CS16115 and CS16140). We would like to thank Prof. CM Lu from IBCAS for the kind donation of RpoB antibody. We appreciate that Prof. XW Sun from SHNU for his help in western blotting. We also thank Prof. Jean-David Rochaix for his help in the language editing of this manuscript. This work was supported by grants from the National Natural Science Foundation of China (31570232 and 31370271) and by the Innovation Program of Shanghai Municipal Education Commission (14YZ066).
Author information
Authors and Affiliations
Corresponding author
Additional information
Qing-Bo Yu and Tuan-Tuan Zhao have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1. Alignment of the amino acid sequences of the pTAC10 orthologs
. The alignment was generated using ClustalW (Thompson et al., 1994) with default parameters and BoxShade 3.21 (http://www.ch.embnet.org/software/BOX_form.html). The positions of identical and similar sequences are indicated in black and grey boxes, respectively. All the sequences used to build the alignment were obtained from NCBI (http://www.ncbi.nlm.nih.gov/). (PDF 581 KB)

Supplementary Figure 2. Identification of phosphorylation sites of pTAC10 by CK2 in vitro
. (A) Phosphorylation sites determined by mass spectrometry are shown. (B) The amino acid sequence around the phosphorylation sites in pTAC10. The phosphoacceptors are indicated by “*”, and the acidic residues at n + 3 are highlighted in grey; (C) An independent phosphorylation analysis of pTAC10. Immunoblot analysis of pTAC10 phosphorylation by cpCK2 in vitro using anti-phospho-Ser, Thr and Tyr antibodies. CK2 was used as positive control. “+” indicates the presence of reagent in the reaction system, and “-” indicates the absence of reagent in the reaction system. “*” indicates the band corresponding to pTAC10. (D) SDS–PAGE analysis of total proteins from unpurified (1) and purified (2) E. coli containing MBP:cpCK2 protein (M) with molecular markers (PageRuler Prestained Protein Ladder, Fermentas) indicated. (JPG 2750 KB)Supplementary material 2 (JPG 843 KB)

Supplementary Figure 3. Mass spectrometric analyses of phosphorylated peptides from of pTAC10 bycpCK2 in vitro
. (A) to (C) LC–MS extracted ion chromatograms showing intensities of the phosphorylated peptide signals from the phosphorylated peptide corresponding to the pTAC10 protein. The phosphorylated Ser residue in the peptide sequence is marked with "ph" on the top of the corresponding amino acid. (JPG 1135 KB)Supplementary material 2 (JPG 843 KB)
Rights and permissions
About this article
Cite this article
Yu, QB., Zhao, TT., Ye, LS. et al. pTAC10, an S1-domain-containing component of the transcriptionally active chromosome complex, is essential for plastid gene expression in Arabidopsis thaliana and is phosphorylated by chloroplast-targeted casein kinase II. Photosynth Res 137, 69–83 (2018). https://doi.org/10.1007/s11120-018-0479-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-018-0479-y