Abstract
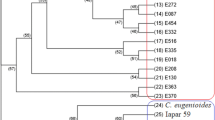
Population genetics analysis is crucial for understanding the degree of linkage disequilibrium in a germplasm collection for association mapping and breeding strategies. Here, we report population structure patterns and genetic differentiation of coffee germplasm collections in China using 19 ISSR markers. A total of 129 unique alleles were detected, with an average of 6.8 alleles per ISSR. The estimated average Nei’s gene diversity (He) index for all tested accessions was 0.49. A clustering pattern correlating with predefined species origin, rather than isolation by distance, was observed for all the tested accessions. C. arabica is highly differentiated from C. liberica and C. canephora, with Fst values of 0.33 and 0.21, respectively. By contrast, C. liberica and C. excelsa Chevalier showed a close genetic association, with the Fst value of 0.04. Significant differentiation was also detected within C. arabica accessions, which were separated in three genetic subgroups supported by pairwise (Fst) index and Nei’s standard genetic distance tests. The Fst outlier test for deviation from expected distribution between Fst and expected heterozygosity identified five putative ISSRs under selection, with two, UBC864 and UBC842, showing signals of positive selection. Our study provides useful information for identifying core collections for genetics studies, breeding programs, and development of future germplasm management strategies.



Similar content being viewed by others
References
Aga E, Bekele E, Bryngelsson T (2005) Inter-simple sequence repeat (ISSR) variation in forest coffee trees (Coffea arabica L.) populations from Ethiopia. Genetica 124:213–221
Antao T, Lopes A, Lopes Beja-pereira A, Luikart G (2008) LOSITAN: workbench to detect molecular adaptation based on a FST-outlier method. BMC Bioinformatic 9:1–5
Beaumont M, Nichols R (1996) Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond Ser B 263:1619–1626
Bland JM, Altman DG (1995) Multiple significance tests: the Bonferroni method. BMJ 310:170
Chaparro A, Cristancho M, Cortina H, Gaitan A (2004) Genetic variability of Coffea arabica L. accessions from Ethiopia evaluated with RAPDs. Genet Resour Crop Evol 51:291–297
Cubry P, Musoli P, Legnaté H, Pot D, de Bellis F, Poncet V, Anthony F, Dufour M, Leroy T (2008) Diversity in coffee assessed with SSR markers: structure of the genus Coffea and perspectives for breeding. Genome 51:50–63
Dieringer D, Schlotterer C (2003) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Draghi J, Parsons T, Wagner G, Plotkin J (2010) Mutational robustness can facilitate adaptation. Nature 463:353–355
Ellstrand N (2014) Is gene flow the most important evolutionary force in plants? Am J Bot 101:737–753
Etienne H, Anthony F, Fernandez S, Lashermes P, Bertrand B (2002) Biotechnological applications for the improvement of coffee (Coffea arabica L.). In Vitro Cell Dev Biol, Plant 38:129–138
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Foll M, Petit R (2009) Genetic consequences of range expansions. Annu Rev Ecol Evol Syst 40:481–501
Fortini L, Schubert O (2017) Beyond exposure, sensitivity and adaptive capacity: a response based ecological framework to assess species climate change vulnerability. Climate Change Responses 4(1)
Freeland J (2006) Molecular ecology. Genetic analysis of multiple populations. John Wiley & Sons, UK
Geleta M, Herrera I, Monzón A, Bryngelsson T (2012) Genetic diversity of Arabica coffee (Coffea arabica L.) in Nicaragua as estimated by simple sequence repeat markers. Sci World J 2012:128–145
Gimase J, Thagana W, Kirubi D, Gichuru E, Gichimu B (2014) Genetic characterization of Arabusta coffee hybrids and their parental genotypes using molecular markers. PCBMB 15:31–42
Greenbaum G, Templeton AR, Bar-David S (2016) Inference and analysis of population structure using genetic data and network theory. Genetics. 202:1299–1312
Hatanaka T, Choi Y, Kusano T, Sano H (1999) Transgenic plants of coffee Coffea canephora from embryogenic callus via Agrobacterium tumefaciens-mediated transformation. Plant Cell Rep 19:106–110
Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, Poncet C, Hochu I, Poirier S, Santoni S, Glémin S, David J (2007) Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol 24:1506–1517
Hyten D, Song Q, Zhu Y, Choi I, Nelson R, Costa J et al (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci U S A 103:16666–16671
International Coffee Council-ICC (2015) Coffee in China. 115th session, Milan, Italy
International Trade Centre-ITC (2010) The coffee sector in China: an overview of production, trade and consumption
Joshi J, Schmid B, Caldeira M, Dimitrakopoulos P, Good J, Harris R, Hector A, Huss-Danell K, Jumpponen A, Minns A, Mulder C (2001) Local adaptation enhances performance of common plant species. Ecol Lett 4:536–544
Kremer A, Ronce O, Robledo-Arnuncio J, Guillaume F, Bohrer G, Nathan R et al (2012) Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol Lett 15:378–392
Lashermes P, Combes M, Robert J, Trouslot P, D'Hont A, Anthony F, Charrier A (1999) Molecular characterization and origin of the Coffea arabica L. genome. Mol Gen Genet 261:259–266
Lashermes P, Andrzejewski S, Bertrand B, Combes MC, Dussert S, Graziosi G, Trouslot P, Anthony F (2000) Molecular analysis of introgressive breeding in coffee (Coffea arabica L.). TAG Theor Appl Genet 100:139–146
Leroy T, Henry A, Royer M et al (2000) Genetically modified coffee plants expressing the Bacillus thuringiensiscry1Ac gene for resistance to leaf miner. Plant Cell Rep 19:382–389
López-Gartner G, Cortina H, Mccouch S, Moncada M (2009) Analysis of genetic structure in a sample of coffee (Coffea arabica L.) using fluorescent SSR markers. Tree Genet Genomes 5:435–446
Maluf MP, Silvestrini M, Ruggiero LMDC, Guerreiro Filho O, Colombo CA (2005) Genetic diversity of cultivated Coffea arabica inbred lines assessed by RAPD, AFLP and SSR marker systems. Sci Agric 62:366–373
Maurin O, Davis A, Chester M, Mvungi E, Jaufeerally-Fakim Y, Fay M (2007) Towards a phylogeny for Coffea (Rubiaceae): identifying well-supported lineages based on nuclear and plastid DNA sequences. Ann Bot 100:1565–1583
Meyer R, Purugganan M (2013) Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet 14:840–852
Mishra M, Slater K (2012) Recent advances in the genetic transformation of coffee. Biotech Res Int 580857:17
Moncada M, Tovar E, Montoya J, González A, Spindel J, McCouch S (2016) A genetic linkage map of coffee (Coffea arabica L.) and QTL for yield, plant height, and bean size. Tree Genet Genomes 12:5
Nagaoka T, Ogihara Y (1997) Applicability of inter-simple sequence repeat polymorphisms in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theor Appl Genet 94:597–602
Ne’de’lec Y et al (2016) Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167:3
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Ogutu C, Fang T, Yan L, Wang L, Huang L, Wang X, Ma B, Deng X, Owiti A, Nyende A, Han Y (2016) Characterization and utilization of microsatellites in the Coffea canephora genome to assess genetic association between wild species in Kenya and cultivated coffee. Tree Genet Genomes 12:54
Park SDE (2001) The Excel microsatellite toolkit (version 3.1). Animal Genomics Laboratory, University College, Dublin
Parter M, Kashtan N, Alon U (2008) Facilitated variation: how evolution learns from past environments to generalize to new environments. PLoS Comput Biol 4:e1000206
Pauls S, Nowak C, Bálint M, Pfenninger M (2013) The impact of global climate change on genetic diversity within populations and species. Mol Ecol 22:925–946
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pearl H, Nagai C, Moore P, Steiger D, Osgood R, Ming R (2004) Construction of a genetic map for Arabica coffee. Theor Appl Genet 108:829–835
Perrier X, Jacquemoud-Collet J (2006) DARwin Software. Montpellier, France: CIRAD. http://darwin.cirad.fr/darwin
Premoli A, Souto C, Allnutt T, Newton A (2001) Effects of population disjunction on isozyme variation in the widespread Pilgerodendron uviferum. Heredity 87:337–343
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond L, Plantegenest M, Vialatte A (2013) Migration and dispersal may drive to high genetic variation and significant genetic mixing: the case of two agriculturally important, continental hoverflies (Episyrphus balteatus and Sphaerophoria scripta). Mol Ecol 22:5329–5339
Rius M, Darling J (2014) How important is intraspecific genetic admixture to the success of colonizing populations. Trends Ecol Evol 29:233–242
Rosenberg N, Burke T, Elo K, Feldmann M, Freidlin P, Groenen M et al (2001) Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics 159:699–713
Scott H, Ryan H, Adi F, Lan Z, Rasmus N, Carlos D (2005) Simultaneous inference of selection and population growth from patterns of variation in the human genome. Evolution 102:7882–7887
Semon M, Nielsen R, Jones M, McCouch S (2005) The population structure of African cultivated rice Oryza Glaberrima (Steud.): evidence for elevated levels of LD caused by admixture with O. sativa and ecological adaptation. Genetics 169:1639–1647
Setotaw T, Caixeta E, Pena G et al (2010) Breeding potential and genetic diversity of BHíbrido do Timor coffee evaluated by molecular markers. Crop Breed Appl Biotechnol 10:298–304
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236:787–792
Sork V (2015) Gene flow and natural selection shape spatial patterns of genes in tree populations: implications for evolutionary processes and applications. Evol Appl 9:291–310
Sousa T, Caixeta E, Alkimim E, de Oliveira A, Pereira A, Sakiyama N, de Resende Júnior M, Zambolim L (2017) Population structure and genetic diversity of coffee progenies derived from Catuaí and Híbrido de Timor revealed by genome-wide SNP marker. Tree Genet Genomes 13:124
Strasburg J, Scotti-Saintagne C, Scotti I, Lai Z, Rieseberg L (2009) Genomic patterns of adaptive divergence between chromosomally differentiated sunflower species. Mol Biol Evol 26:1341–1355
Tanksley S, McCouch S (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Turner T, Hahn M (2010) Genomic islands of speciation or genomic islands and speciation. Mol Ecol 19:848–850
Van der Vossen H (1985) Coffee breeding and selection. In: Clifford MN, Wilson RC (eds) Coffee: botany, biochemistry and production of beansand beverages. Croom Helm, London, New York, Sydney, pp 48–97
Vignuzzi M, Stone J, Arnold J, Cameron C, Andino R (2006) Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348
Wakeley J (2000) The effects of subdivision on the genetic divergence of populations and species. Evolution 54:1092–1101
Wakeley J, King L, Wilton P (2016) Effects of the population pedigree on genetic signatures of historical demographic events. Proc Natl Acad Sci U S A 113:7994–8001
Weir BS, Cockerham C (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Williamson S, Hernandez R, Fledel-Alon A, Zhu L, Nielsen R, Bustamante C (2005) Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc Natl Acad Sci U S A 102:7882–7887
Yapo A, Louarn J, Montagnon C (2003) Genetic gains for yield after two back-crosses of the interspecific hybrid Libusta (Coffea canephora P. x C. liberica Bull. Ex Hiern) to C. canephora P. Plant Breed 122:288–290
Zhang HL, Zhou J, Chen H, Song Z, Peng G, Pereira Z, Silva A, Varzea MV (2013) Arabica coffee production in the Yunnan Province of China. Conference: Proceedings of the 24th International Conference on Coffee Science (ASIC)
Funding
This project was supported by funds received from Hainan Provincial Natural Science Foundation of China (No.2018CXTD342), Overseas Construction Plan for Science and Education Base, China-Africa Center for Research and Education, Chinese Academy of Sciences (Grant No. SAJC201327), and Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No.1630142018001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Data Archiving Statement
Primer sequences of the ISSR markers are provided in Table 1.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOC 398 kb)
Rights and permissions
About this article
Cite this article
Yan, L., Ogutu, C., Huang, L. et al. Genetic Diversity and Population Structure of Coffee Germplasm Collections in China Revealed by ISSR Markers. Plant Mol Biol Rep 37, 204–213 (2019). https://doi.org/10.1007/s11105-019-01148-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-019-01148-3