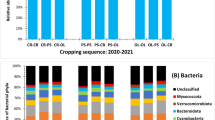
Abstract
Aims
Saprotrophic soil fungi participate in biomass mineralization, inhibit pathogen development and promote plant growth. Pathogens accumulate in soil and decrease crop yields. The structure of fungal communities is determined mainly by the organic matter content and pH of soil. Little is known about the influence of crop rotation and long-term monoculture on saprotrophic fungi that decompose plant roots and crop residues as sources of soil biomass.
Methods
Fungal communities that promote plant growth (arbuscular mycorrhizal fungi (AMF), yeasts, Trichoderma spp.), cellulolytic fungi and pathogenic species were analyzed in a 6-year crop rotation system (maize – spring barley – peas – winter rapeseed – winter wheat – sugar beets) and in 50-year maize monoculture. Fungal DNA was extracted from the rhizosphere and plant roots, and the ITS2 region of fungal rDNA was analyzed by high-throughput sequencing. In both treatments, weeds were controlled chemically (terbuthylazine + mesotrione + s-metolachlor) or mechanically.
Results
A total of 311 fungal species were identified. The biodiversity of soil fungi, in particular AMF and yeasts, was higher in monoculture than in crop rotation. Maize pathogens were more frequently identified in monoculture, whereas species of the genus Trichoderma were more prevalent in crop rotation. Herbicides clearly increased the abundance of cellulolytic fungi of the phyla Mucoromycota and Mortierellomycota, Mortierella spp. and Minimedusa polyspora. The abiotic properties of soil were affected by the cropping sequence. The content of organic carbon (Corg) and the availability of P and Mg decreased in monoculture. Maize yields were bound by a strong positive correlation with the availability of macronutrients and Corg in soil, as well as a weak positive correlation with the abundance of Trichoderma spp., Mucoromycota and Mortierellomycota.
Conclusions
Fungi exert a complex and ambiguous effect on maize biomass yields, whereas a decrease in the macronutrient content of soil in monoculture strongly decreases maize yields. In the long term, the cropping sequence considerably influences the structure of the soil microbiome which can be a reservoir of unique species and species that minimize the negative effects of monoculture in agroecosystems.
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Plants and soil are increasingly described as reservoirs of bacterial and fungal microbiomes (Gałązka and Grządziel 2018; Duniere et al. 2017; Fadiji et al. 2020) whose structure can affect plant development (Guler et al. 2016; Kusuma et al. 2017; Sobowale et al. 2010). Soil microbial communities are very important components of the ecosystem. According to estimates, 1 g of soil is colonized by around 109 microorganisms (Delmont et al. 2021; Kong et al. 2020). Fungal communities play diverse roles in the soil environment, and some fungal species infect the roots and aerial parts of maize plants. Soil can be a reservoir of mycotoxin-producing Fusarium graminearum and F. verticillioides, which are among the most dangerous pathogens of maize (Pechanova and Pechan 2015; van Capelle et al. 2021). Many soil fungi, including Sporotrichum pruinosum and Stachybotrys atra, participate in the decomposition of plant residues and nutrient cycling, thus increasing the productivity of agricultural soils (Mahmood et al. 1986). In a study by Kusuma et al. (2017), fungi of the genera Trichoderma and Penicillium inhibited the development of Rhizoctonia solani which causes banded leaf and sheath blight in maize. Most agricultural treatments significantly affect soil structure as well as the abundance and structure of fungal communities (Beauregard et al. 2013; Sasvári et al. 2011; Wang et al. 2017; Fadiji et al. 2020). The substances secreted by the roots of several maize varieties exerted varied effects on fungal communities (Kong et al. 2020). In a study by Tyagi et al. (2018), herbicides significantly reduced fungal populations in soil under maize cultivation.
Maize is one of the most important food crops around the world, and its global production reached 1162.4 million tons in 2020 (FAOSTAT 2022). The United States, China and Brazil are the leading producers of maize. These countries supply nearly 60% of maize grain on the global market, whereas Poland produces around 6 million tons of maize grain each year. Maize is also grown for silage which is used as cattle feed. The United States is also the leading producer in this category, and the area under maize cultivated for silage accounts for 20% of the total area. In the European Union (EU 27), maize is grown for silage on around 6,3 million hectares each year. Germany and France are the leading producers of maize harvested for silage, whereas Poland ranks third in the EU with an area of 675 thousand hectares (EUROSTAT 2022).
Monoculture, namely the practice of growing the same crop species in the same field year after year, is relatively popular because it enables farmers to specialize in a particular crop and maximizes profits (Gałązka and Grządziel 2018; Woźniak 2019). However, monoculture decreases crop yields already after several years, mainly due to the depletion of soil nutrients (Mao et al. 2021). Long-term monoculture affects the structure of soil microbial communities, for example, by promoting the growth of selected fungal pathogens of crops Fusarium oxysporum and F. solani (Liu et al. 2019; Strom et al. 2020), but it does not significantly decrease the diversity of mycorrhizal fungi (Glomus spp.) colonizing maize roots (Sasvári et al. 2011). Soil-dwelling microorganisms can mobilize nutrients (such as nitrogen-fixing bacteria, Kosakonia sacchari, Bloch et al. 2020) and enlarge the root absorption area (as the mycorrhizal fungus Rhizoglomus intraradices; Smith and Smith 2012); therefore, the bioavailability of nutrients can be limited in the absence of specific microbial species in soil (Mao et al. 2021; Bergmann et al. 2020; Roumet et al. 2016).
Next-generation sequencing methods support reliable identification of microbial species and quantitative changes in their populations (Fadiji and Babalola 2020). The internal transcribed spacer (ITS) region of fungal DNA has been sequenced to study the composition and diversity of endophytic fungi in crops, especially maize (Fadiji et al. 2020). In the present study, next-generation sequencing was applied to compare the structure of fungal communities, maize biomass yields. The content of organic matter and minerals was also analyzed in soil under maize grown in 50-year monoculture and crop rotation systems, with and without the use of herbicides, in order to investigate the interactions between maize yields, soil fungi, and nutrient concentrations in soil. The aim of this study was to check the following hypotheses: (1) monoculture strongly influences the structure of fungal communities in soil and induces significant changes in the abundance of non-pathogenic fungal functional groups (mycorrhizal fungi that suppress plant pathogens and promote the decomposition of organic compounds); (2) pathogens that accumulate in soil inhibit the growth of maize plants in monoculture; (3) herbicides increase the abundance of fungi that decompose the organic matter of damaged weeds. The aim of this study was to evaluate quantitative changes in the structure of fungal communities colonizing the rhizosphere in maize monoculture, including under the influence of herbicides, to examine the interactions between fungal taxonomic and functional groups vs. maize yields, and to determine nutrient concentrations in soil.
Materials and methods
Site description, weather conditions, agricultural practices and sampling
The experimental material originated from a long-term maize monoculture experiment established in 1968 in Bałcyny (53°35ʹ49ʺ N, 19°51ʹ20ʺ E; Poland). The analyzed region is characterized by a moderate climate with mean annual temperature of 7.1 °C and mean annual precipitation of around 600 mm (Kuchar et al. 2021). Maize biomass was collected between 2017 and 2019, and weather conditions for this period are presented in Table 1. Weather conditions were analyzed with the use of the Selyaninov hydrothermal coefficient, calculated with the formula K = P/0.1·Et, where K is the hydrothermal coefficient, P – total monthly precipitation, and Et – sum of mean daily air temperatures in a given month. In 2017, precipitation was high between June and September when maize is harvested (K = 2.31). 2018 was a rather dry year (K = 1.05). Following a dry spring (April–May), which marked the beginning of the maize growing season, precipitation increased in June and July, whereas rainfall deficit was noted in August and September. In 2019, April was an extremely dry month (K = 0.01), but the remaining months of the growing season were characterized by favorable hydrothermal conditions for the growth and development of maize plants.
The field experiment had a randomized block design, where treatments were randomly assigned to sub-blocks in four replications. Plot area was 1.8 × 14 m. Maize was grown in a 6-crop rotation system: maize – spring barley – peas – winter rapeseed – winter wheat – sugar beets (treatments: CR – without herbicides, and CRH – with herbicides), and in a 50-year monoculture system (treatments: M – without herbicides, and MH – with herbicides). Crops were grown in a conventional tillage system. Medium-maturing maize variety Lg 31.233, a three-way hybrid recommended for silage production, was sown between 2017 and 2019. The optimal dates of agricultural treatments in maize production are presented in Table 2. Each year, the health status of plants was monitored twice during the growing season, in full flowering (BBCH 65) and medium milk (BBCH 75) stages (Meier 1997).
In crop rotation (CRH) and in monoculture (MH), weeds were controlled after emergence with Lumax 537.5 SE (terbuthylazine; triazine herbicide) – 187.5 g l–1 (16.9%), mesotrione (triketone herbicide) – 37.5 g l–1 (3.39%), and s-metolachlor (chloroacetanilide herbicide) – 312.5 g l–1 (28.2%)) at 3.5 L ha–1 (Syngenta, Poland) on the dates indicated in Table 2. In treatments not protected with herbicides (CR and M), weeds were controlled mechanically, and weed residues were left on the soil surface.
In 2017–2019, mineral fertilizers were applied at identical rates in all treatments (CR, CRH, M and MN): N – 120 kg ha–1 (N1 – 60 kg + N2 – 60 kg), P – 70 kg ha–1, and K – 150 kg ha–1, on the dates indicated in Table 2. In monoculture, crop residues had been recycled in the field since 1968. Maize stem and root residues were incorporated into the soil during tillage, and manure was applied every three years at 15 t ha–1. In crop rotation, crop residues were incorporated into the soil during tillage, and manure was applied to sugar beets every six years at 30 t ha–1 (last application in 2016). Maize green forage (biomass) was harvested in the milk/dough stage (BBCH 86). The biomass yield per plot was determined by weighting immediately after harvest.
Soil samples for analyses of the fungal microbiome were collected on 10 July (BBCH 65) 2018. Three maize root samples (with soil) were collected at a depth of 20 cm in three random locations in each treatment with the use of a soil probe (length – 20 cm, internal diameter – 16 mm). The collected samples had a size of approximately 16 mm × 5 cm.
Agronomic characteristics of soil
The field experiment was established in a slightly undulating area, on haplic Luvisols formed from silty light loam (IUSS Working Group WRB 2015). Soil samples were analyzed with the use of the method described by van Reeuwijk (2002). Soil pH was determined potentiometrically in water and potassium chloride solution (1 M KCl dm–3). Total organic carbon (Corg) was measured with a Vario Max Cube CN elemental analyzer. In the arable layer, the content of clay, silt, and sand was 2–4%, 26–39% and 57–72%, respectively (Rychcik et al. 2006). In crop rotation and monoculture, soil was slightly acidic; the content of Corg was determined at 10.25 and 7.23 g kg–1; the content of available phosphorus was very high, and the content of available potassium was moderate. The content of available magnesium was moderate in crop rotation and low in monoculture (Table 3).
DNA isolation from soil
DNA was isolated directly from the rhizosphere and plant roots with the Soil DNA Purification Kit (EURx LTD, Poland) according to the manufacturer’s instructions. The samples were homogenized mechanically in bead tubes with the use of the Star Beater for Molecular Biology (Bio-Strategy Ltd., USA). The quantity and quality of the isolated DNA were checked by measuring absorption at a wavelength of 260 nm and 280 nm (NanoDrop 2000, Thermo Scientific, Polska). DNA was stored at a temperature of -20 °C.
Amplification of DNA fragments by PCR
The metagenomic sequencing analysis of the hypervariable ITS2 region in fungi was conducted according to a previously described method (Jastrzębska et al. 2020). The selected region was amplified and the library was prepared with the use of fungi-specific primers fITS7 (GTGARTCATCGAATCTTTG) and ITS4 (TCCTCCGCTTATTGATATGC), supplemented with an overhang adapter sequence at the 5’ end of each primer. PCR was conducted with the use of the Q5 Hot Start High-Fidelity 2X Master Mix. Dual-indexed libraries were prepared with the Nextera XT Index Kit.
Illumina MiSeq sequencing
Paired-end (PE) DNA sequencing was performed in the MiSeq sequencer (Illumina, Polska) (2 × 250 bp) with the Illumina Kit v2 (Genomed 2020, Poland). The sequencing procedure was described previously by Jastrzębska et al. (2020). Fungal DNA samples were sequenced by Genomed, a biotechnology company (www.genomed.pl). Operational taxonomic units (OTUs), at a 97% similarity threshold, were determined using the simple alignment search tool (BLAST).
Statistical analysis
Data were processed by analysis of variance (ANOVA), linear correlation analysis and principal component analysis (PCA) in the Statistica 13 program (StatSoft 2019). The significance of differences between means was evaluated by Duncan’s test (p < 0.01). The Shannon–Wiener diversity index (Hw) was calculated from the equation Hw = − ∑ pi(ln pi), where pi is the proportion of individuals found in the ith OTU. The similarities between fungal communities in the analyzed samples were determined based on normalized Euclidean distances. Spearman’s correlation coefficients between fungal taxonomic and functional groups vs. maize yields and nutrient concentrations in soil were calculated.
Results
Fungal community profiles
A comparison of fungal community profiles in samples of rhizosphere soil from monoculture and crop rotation systems revealed that the vast majority of OTUs were represented by the phyla Ascomycota (75.64% on average) and Basidiomycota (14.67%) (Table 4). The reads associated with the phylum Mortierellomycota accounted for nearly 8.07% of total OTUs. The remaining reads were linked with the kingdoms Aphelidiomycota (0.03%), Chytridiomycota (0.83%), Glomeromycota (0.54%), Mucoromycota (0.22%) and Olpidiomycota (0.006%) that occurred sporadically and accounted for less than 2% of total OTUs.
The influence of crop rotation and herbicide treatment on fungal community profiles
A total of 423,297 high-quality ITS2 sequences were obtained from all samples (Table 4). When grouped at 97% sequence similarity, a total of 311 species were identified in all treatments, including 243, 204, 172, and 198 species in treatments M, MH, CR, and CRH, respectively. The fungal community from rhizosphere soil in monoculture (M) was characterized by greater biodiversity than the community from crop rotation (CR), and the values of the Shannon–Wiener diversity index reached 1.465 and 1.217, respectively. The application of herbicides decreased fungal community biodiversity in monoculture (Hw = 1.389), but it had no apparent effect in treatment CRH (Hw = 1.254).
In all soil samples, the dominant fungal phyla were Ascomycota (relative OTU abundance from 57.45% in CRH to 92.22% in CR), Basidiomycota (5.12% in CR to 30.36% in CRH) and Mortierellomycota (2.34% in CR to 11.69% in MH). Rhizosphere soil from CR treatments was far less abundant in taxa belonging to the phylum Ascomycota than the phyla Basidiomycota and Mortierellomycota. The abundance of OTUs originating from the phyla Chytridiomycota and Glomeromycota was highest in monoculture (M), although it was relatively low at only 1.73% and 1.53%, respectively. The relative abundance of OTUs associated with Aphelidiomycota, Mucoromycota and Olpidiomycota did not exceed 0.26% (Table 4).
The following percentage of dataset reads was classified at kingdom, phylum, class, order, family, genus and species level: 100%, 98.40%, 85.19%, 84.63%, 74.59%, 69.29% and 61% for M; 100%, 98.89%, 89.03%, 88.76%, 76.02%, 73.69% and 66.07% for MH; 100%, 98.99%, 90.34%, 89.87%, 86,60%, 77,85% and 76.96% for CR; and 100%, 98.42%, 87.47%, 85.85%, 81.38%, 76.16% and 68.29% for CRH, respectively (Table S1).
In the total number of 311 identified species of the kingdom Fungi, the abundance of OTUs associated with 36 dominant species was determined at 55.01% in M, 61.47% in MH, 73.49% in CR, and 62.33% in CRH. Five of the dominant species formed a group of hyperdominant species with a minimum total number of 15,700 OTUs in all treatments. These were: Trichoderma hamatum, Didymella sancta, Talaromyces sayulitensis, Solicoccozyma fuscescens, and Minimedusa polyspora (Fig. 1). Trichoderma hamatum dominated in CR (25,215 OTUs), M. polyspora in CRH (24,644 OTUs), and D. sancta in M and MH (12,667 OTUs). An analysis of the abundance of the dominant fungal species in treatments revealed similarities between treatments M and MH and the absence of similarities between treatments CR and CRH (Fig. 1).
Species of the phylum Ascomycota were classified into 24 orders, 64 families and 124 genera, where 16 genera contained three or more species each, and 12 genera contained two species each (Table S2). Species of the phylum Basidiomycota were classified into 23 orders, 41 families and 76 genera, where 5 genera contained three or more species each, and 16 genera contained two species each (Table S3).
The influence of crop rotation on the number of fungal taxonomic and functional groups
The identified fungal species were divided into six taxonomic or functional groups: (1) pathogens, (2) yeasts, (3) Glomeromycota, (4) Trichoderma spp., (5) Mucoromycota + Mortierellomycota, and (6) Eurotiales. The relative abundance of Glomeromycota, Mucoromycota, Mortierellomycota, Eurotiales and yeasts was 50-, 2-, 23- and 1.5-fold higher, respectively, in soil samples from monoculture (M) than crop rotation (CR) (Fig. 2). Crop rotation increased the abundance of all pathogens 1.7-fold and the relative abundance of Trichoderma spp. tenfold. Herbicide application (MH and CRH) clearly decreased the abundance of pathogens and increased the abundance of Mucoromycota and Mortierellomycota, compared with treatments without herbicides (M and CR).
The identified fungal species were divided into four taxonomic and functional groups: (1) pathogens and Glomeromycota, (2) yeasts and Eurotiales (3) Trichoderma spp., and (4) Mucoromycota and Mortierellomycota (Fig. 3). Two clades were identified in the first pattern (M, MH and CR, CRH). Three clades were identified in the remaining patterns. In the group of yeasts, Trichoderma spp. and Eurotiales, the first clade comprised M and MH, the second – CR, and the third – CRH. The second and third clades differed in similarity to the first clade. The fourth clade (Mucoromycota and Mortierellomycota) was unique because the fungal community in M was grouped with CR.
Structure of selected fungal taxonomic and functional groups
Fungal pathogens accounted for 22.74% of total OTUs, and they were represented by 19 species, including several species that are potentially pathogenic for maize: Ustilago maydis, Nigrospora oryzae and Gibberella avenacea (Fig. 4). Ustilago maydis, a biotrophic pathogen of maize, occurred only in monoculture (3.57% of total OTUs in M). Gibberella avenacea (anamorph of Fusarium avenaceum), the dominant pathogen of the genus Fusarium, was identified mainly in CR. Polyphagous pathogens of the genus Didymella that target other crop species were dominant in treatments M (47.93%) and MH (40.28%). The soybean pathogen Boeremia exigua was noted mainly in treatment CRH, and it accounted for 34.46% of total OTUs in CRH.
Yeasts accounted for 5.32% of total OTUs. Twenty-eight yeast species were identified in treatments M and MH, and 24 species – in CR and CRH (Fig. 5). The dominant species Solicoccozyma fuscescens accounted for 62.49%, 81.28%, 55.72% and 65.54% of total OTUs in M, MH, CR and CRH, respectively. Most yeast species were more abundant in monoculture than in crop rotation. The greatest differences were observed in the abundance of S. fuscescens, Naganishia vaughanmartiniae, Rhodotorula glutinis and Malassezia restricta.
Only four Glomeromycota species were identified, and the remaining OTUs were identified to genus, family, order or phylum level (Fig. 6). Glomeromycota accounted for only 0.54% of total OTUs. The abundance of OTUs associated with most of the identified taxa, including Glomus spp., Paraglomus spp., Ambispora fennica, Claroideoglomus claroideum and Funneliformis mosseae, was higher in treatments M and MH than in CR and CRH.
Fungi of the genus Trichoderma accounted for 11.3% of total OTUs (Fig. 7). Nine Trichoderma species were identified, and T. hamatum was the hyperdominant species that accounted for 96.18%, 96.86%, 71.53% and 37.49% of total OTUs in treatments M, MH, CR and CRH, respectively. Due to the high abundance of T. spirale, T. lanuginosum and T. harzianum in crop rotation, in particular in treatment CR, the percentage share of Trichoderma spp. was higher than in monoculture.
Species belonging to the phylum Mortielellomycota accounted for 4.63% of total OTUs, whereas species belonging to the phylum Mucoromycota accounted for only 0.22% of total OTUs (Fig. 8). The three dominant species, Mortierella exigua, M. elongata and M. alpina, were identified mainly in herbicide-treated plots, where the abundance of OTUs associated with M. exigua was up to 62.6 times higher in monoculture (MH/M) and 31.8 times higher in crop rotation (CRH/CR).
A total of 16 species belonging to the order Eurotiales were identified and they accounted for 7.02% of total OTUs (Fig. 9). Talaromyces sayulitensis was the dominant species in all treatments, accounting for 69.22%, 76.04%, 53.04% and 90.57% of OTUs in M, MH, CR and CRH, respectively. Penicillium jensenii, P. rolfsii, P. simplicissimum, and P. spathulatum were also highly abundant and dominated in monoculture soil.
Maize yields
During the entire study, disease symptoms and pests were noted only sporadically in maize plants grown in crop rotation and monoculture. In 2017–2019, the average yield of maize biomass was 23.8% lower in monoculture than in crop rotation (Table 5). Herbicide treatment increased yields by an average of 15.2% in monoculture and 17.5% in crop rotation. The highest yields in monoculture (74.2 t ha–1) and in crop rotation (100.8 t ha–1) were noted in 2017, which can probably be attributed to optimal precipitation levels during the growing season (Tables 1 and 5).
Principal component analysis
In the principal component analysis based on 12 maize and soil parameters (maize yields, abiotic and biotic soil properties) in monoculture and crop rotation systems with and without the use of herbicides, the first two principal components explained 77.87% of total variance (Fig. 10). The first principal component (PC1) explained 64.24% of total variance, and it was dominated by yield, content of available K, and the abundance of pathogenic fungi that can indirectly positively influence maize yields: Trichoderma spp., Mucor spp. and Mortierella spp. The second principal component (PC2) explained 23.63% of total variance, and it was composed mainly of nutrient availability in soil and the abundance of fungal communities that promote plant growth (Eurotiales, Glomeromycota, yeasts). Nutrient concentrations were grouped around maize yields.
Several significant negative correlations were observed between fungal abundance and the abiotic properties of soil (Table S4, Fig. 11). The abundance of Eurotiales and Glomeromycota was negatively correlated with the content of available Mg (r = -0.966) and soil pH (r = -0.973), respectively.
Graphical heat map representation of correlation matrix between selected fungal groups, nutrient concentrations in soil, and maize yields. Y 2017—yield in 2017, Y 2018—yield in 2018, Y 2019—yield in 2019, Y (mean)—mean of yield in 2017–2029, pH—soil pH in KCl, Corg—soil organic carbon, P—phosphorus available, K—potassium available, Mg—magnesium available, S-W index—Shannon–Wiener index, Euro—Eurotiales, Glom—Glomeromycota, Mort, Muc—Mortierellomycota, Mucoromycota, Trich—Trichoderma, Path—Pathogens, U. may—Ustilago maydis, N. ory—Nigrospora oryzae, Drech—Drechslera spp., G. ave—Gibberella avenacea, S. ter—Setophoma terrestris, P. ave—Parastagonospora avenae, Alt—Alternaria spp., Did—Didymella spp., Fus – Fusarium spp., F. aqu—Fusicolla aquaeductuum, B. exi—Boeremia exigua, C. glo—Colletotrichum gloeosporioides
Discussion
The abundance of pathogenic and non-pathogenic rhizosphere fungi in maize monoculture established in 1968 differed considerably from that noted in a 6-year crop rotation system, which could explain the observed difference in maize yields. The present findings corroborate the results of previous studies where the abundance of fungal OTUs in soil under maize cultivation was relatively high in both crop rotation and in long-term monoculture (Sasvári et al. 2011; Kong et al. 2020). In the current study, the biodiversity of the fungal community was very high in monoculture, and it decreased in crop rotation and under the influence of herbicide treatment. Moreno et al. (2021) also demonstrated that agricultural treatments that reduce soil moisture content can decrease the diversity of the fungal microbiome.
In the present study, the relative abundance of all pathogens was high, and it accounted for around 24% of total OTUs. Similar observations were made by Mao et al. (2021), in whose study, potentially pathogenic fungi accounted for 18% of the fungal community. Ustilago maydis, a biotrophic pathogen that causes corn smut (González-Prieto et al. 2014), was identified only in monoculture. Corn smut was observed sporadically, and its severity did not differ between treatments. The low prevalence of corn smut can be probably attributed to the fact that the tested maize varieties were characterized by increased resistance to U. maydis infections (Pathi et al. 2020).
In this study, negative and non-significant correlations were found between maize yields and the abundance of hemibiotrophic and necrotrophic pathogens of maize, including Drechslera spp. (Sugawara et al. 1987), Nigrospora oryzae (Standen 1945), Gibberella avenacea (Naef and Défago 2006), Setophoma terrestris (Lević et al. 2013) and Parastagonospora avenae (Marin-Felix et al. 2017). Similar observations were made by Mao et al. (2021) who analyzed maize monoculture. In the present study, pathogens that target crop species other than maize were also identified in soil, including Boeremia exigua, Colletotrichum gloeosporioides and Didymella santa. These pathogens were most abundant in crop rotation, in particular in the treatment without herbicides (CR). Their abundance was not negatively correlated with maize yields.
Colletotrichum gloeosporioides infections have been reported in several dozen plant species, including coffee, cereals, mangoes, strawberries and apples (Huang et al. 2019). In the group of pathogens that do not target maize, only Didymella spp. was bound by a non-significant negative correlation with maize biomass yields. Didymella is one of the 31 genera of the family Didymellaceae that includes cosmopolitan polyphagous pathogens damaging the stems and leaves of plants growing in diverse environments (Keirnan et al. 2021). Didymella sancta (syn. Phoma sancta), a dominant species in the current study, has been found to infect Ailanthus altissima in South Africa and Opuntia ficus-indica in Argentina (Aveskamp et al. 2009; Chen et al. 2017; Keirnan et al. 2021). Phoma species colonize soil and a wide range of host plants as primary pathogens, opportunistic pathogens, saprobionts or endophytes (Aveskamp 2014).
In this study, several potentially beneficial fungi were identified in monoculture soil, including seven genera of the phylum Glomeromycota. This phylum comprises species of arbuscular mycorrhizal fungi (AMF). In contrast, only one genus of the Glomeromycota, i.e. Paraglomus, was identified in crop rotation. Mwakilili et al. (2021) demonstrated that the genus Glomus was enriched in maize monoculture plots. The abundance of Glomeromycota fungi was significantly negatively correlated with soil pH which was lower in monoculture. According to Beauregard et al. (2013), soil pH as well as phosphorus fertilization rates can influence AMF abundance. In the cited studies, higher rates of nitrogen and phosphorus fertilizers clearly suppressed the growth of AMF. Research shows that plant roots that are initially exposed to mycorrhizal fungi are increasingly colonized by AMF during the growing season (Henry and Kosola 1999), and crops sown at the beginning of the growing season are repeatedly colonized each spring (Beauregard et al. 2013). However, the cited authors observed that plant roots can be colonized by both newly emerged fungi as well as AMF that had survived on degraded roots from previous years. These observations suggest that root residues in monoculture were less degraded due to a smaller number of cellulolytic fungi, which, consequently, increased the abundance of AMF. Johnson et al. (2003) postulated that roots recruit only a small number of AMF taxa that survive in the soil as spores. In the current study, Ambispora fennica was detected only in monoculture. In the present study, the genus Glomus was noted only in monoculture soil in the treatment without herbicides, and its abundance was determined at 26 OTUs. Our results stand in contrast to the findings of other authors. Beauregard et al. (2013) reported on the high abundance of G. fasciculatum, G. intraradices and G. irregulare (currently Rhizoglomus) in soil under maize cultivation, whereas Bhadalung et al. (2005) detected five unidentified species of the genus Glomus. The lower abundance of Glomus spp. in this study can be probably attributed to different sampling dates. Soil samples were collected in July, when the abundance of Glomus fungi is much lower than in October, November and December (Bhadalung et al. 2005).
Other AMF species, including Claroideoglomus claroideum and Funneliformis mosseae, were also detected in monoculture (M and MH). Funneliformis caledonium was identified only in the herbicide treatment in crop rotation (CRH). According to Tanwar et al. (2013), maize, lemon grass (Cymbopogon nardus (L.) Rendle) and palmarosa are the preferred host species for F. mosseae. In a study by Malicka et al. (2021), Funneliformis caledonium was most tolerant to phenol and PAHs and was characterized by the highest potential for plant growth promotion. In the work of Wang et al. (2021), root inoculation with F. caledonium increased maize yields, foliar content of jasmonic acid and salicylic acid, foliar content of Bt toxin, and Bt gene expression in the leaves of Bt maize.
In the current study, several dozen yeast species, Trichoderma spp. and Penicillium spp. of the order Eurotiales were identified in soil under maize and accounted for 33.64% of total OTUs. Nine species of the genus Trichoderma were detected mainly in crop rotation. Trichoderma species are known for their ability to suppress maize pathogens. Trichoderma hamatum and T. longibrachiatum inhibited the development of Fusarium verticillioides colonies in vitro (Sobowale et al. 2010), whereas T. longibrachiatum and T. asperelloides suppressed the growth of the maize pathogen Magnaporthiopsis maydis in vitro, in the greenhouse and in the field (Degoni and Dor 2021). In turn, the T. atroviride BC0584 isolate enhanced the emergence of maize seedlings in the greenhouse and in the field (Coninck et al. 2020). In the present study, the higher abundance of Trichoderma OTUs in crop rotation probably contributed to maize growth by eliciting plant defense mechanisms (Coninck et al. 2020; Guler et al. 2016), and exerting antagonistic effects on plant pathogens (Narayanasamy 2013).
Talaromyces sayulitensis, a predominant species in the present study, had been previously identified in maize grain (Ekpakpale et al. 2021), shale by-products and household dust (de Goes et al. 2017). In the present study, Talaromyces sayulitensis and other Penicillium spp. were identified mainly in monoculture. Penicillium species produce antibiotics and secondary metabolites with biological activity (Saveetha et al. 2021). According to the cited authors, the accumulation of Penicillium fungi in maize roots could have contributed to higher yields.
Many of the yeast species identified in this study are widely used in biotechnological processes (Nandya and Srivastavab 2018), biological crop protection (Köhl et al. 2019) and food production (El-Ghwas et al. 2014). These fungi were detected mainly in monoculture, and one of the 35 identified species, Candida sake (widely described in the literature), is used to protect grapes against storage pathogens (Calvo-Garrido et al. 2014). Debaryomyces hansenii suppressed Monilinia fructicola in apple fruit (Czarnecka et al. 2019) and Fusarium spp. in wheat (Wachowska et al. 2020). Rhodotorula glutinis inhibited the development of Botrytis cinerea (Li et al. 2016), Sporobolomyces roseus reduced the prevalence of blue mold in apples (Janisiewicz et al. 1994), and Papiliotrema flavescens decreases the severity of Fusarium head blight (Schisler et. al. 2019). Rhynchogastrema coronatum, also noted in the present study, acts as a hyperparasite of fungi (Liu et al. 2015). Solicoccozyma fuscescens, the most frequently identified yeast species in this study, had been previously detected in agricultural soil (Mašínová et al. 2017), but its ecosystem functions have not yet been fully elucidated. To the best of our knowledge, several yeast species identified in the current study had not been previously identified in soil samples in Central Europe. These include Saitozyma paraflava, the species from Thailand and Japan, and Filobasidium oeirense which was the dominant species in durum wheat grain in Italy.
In the present study, the dominant fungal species Minimedusa polyspora was highly abundant in the herbicide treatment in crop rotation, which could be attributed to the presence of sugar beet residues that are rich in sucrose as well as weed residues. According to Pinzari et al. (2017), M. polyspora is a highly efficient pioneer colonizer that requires little nitrogen, is characterized by rapid growth, adapts its metabolism to substrate modifications, and secretes substances that inhibit the development of competitive species. The cited study demonstrated that M. polyspora has a preference to polysaccharides in early stages of growth but prefers hexoses and then oligosaccharides in later phases of development. Pinzari et al. (2017) found that M. polyspora was capable for concentrating several important biogenic elements (N, P, S, K and Ca). Fungi of the phyla Mucoromycota and Mortierellomycota, which were most abundant in herbicide treatments, probably had similar properties.
In this study, herbicide application in monoculture decreased the biodiversity of the fungal community relative to the treatment without herbicides. The above resulted mainly from the fact that fungal species representing the functional group of fungi with cellulolytic properties participated in the decomposition of weed organic matter. Three dominant species of the genus Mortierella (M. exigua, M. elongata and M. alpina) were overabundant in the monoculture treatment with herbicides. In a recent review article (Ozimek and Hanaka 2021), species of the genus Mortierella were classified as plant growth-promoting fungi (PGPF) that increase the availability of P and Fe in soil, and are capable of synthesizing phytohormones and 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase. These species can survive in highly unsupportive environments by relying on sources of carbon in polymers such as cellulose, hemicellulose and chitin, which makes them highly effective plant growth modifiers in agricultural production (Ozimek and Hanaka 2021).
Summary
The biodiversity of soil fungal communities, mainly AMF and yeasts, was higher in monoculture than in crop rotation. Biomass-decomposing fungi of the phyla Mucoromycota and Mortierellomycota as well as Minimedusa polyspora partly minimized the negative effects of monoculture farming. Species of the genus Mortierella additionally promoted the growth of maize plants. Lower maize yields in monoculture can be attributed to the presence of root pathogens and soil nutrient depletion. The soil microbiome is influenced by numerous factors, including the complex interactions between soil abiotic properties and plant root characteristics. Further research is needed to explore the ecological functions of fungal communities in agricultural soils, including the role of biomass-decomposing species and yeasts in promoting plant growth.
Data availability
The datasets generated during the current study are available from the first author on reasonable request.
References
Aveskamp M (2014) Phylogeny and DNA-based identification in Phoma and related genera. PhD thesis Wageningen University, Wageningen, pp 206
Aveskamp MM, Verkley GJ, de Gruyter J, Murace MA, Perello A, Woudenberg JH, Groenewald JZ, Crous PW (2009) DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 101(3):363–382. https://doi.org/10.3852/08-199
Beauregard MS, Gauthier MP, Hamel C, Zhang T, Welacky T, Tan CS, St-Arnaud M (2013) Various forms of organic and inorganic P fertilizers did not negatively affect soil- and root-inhabiting AM fungi in a maize–soybean rotation system. Mycorrhiza 23:143–154. https://doi.org/10.1007/s00572-012-0459-6
Bergmann J, Weigelt A, van der Plas F, Laughlin DC, Kuyper TW, Guerrero-Ramirez N, Valverde-Barrantes OJ, Bruelheide H, Freschet GT, Iversen CM, Kattge J, McCormack ML, Meier IC, Rillig MC, Roumet C, Semchenko M, Sweeney CJ, van Ruijven J, York LM, Mommer L (2020) The fungal collaboration gradient dominates the root economics space in plants. Scie Advances 6(27):eaba3756. https://doi.org/10.1126/sciadv.aba3756
Bhadalung NN, Suwanarit A, Dell B, Nopamornbodi O, Thamchaipenet A, Rungchuang J (2005) Effects of long-term NP-fertilization on abundance and diversity of arbuscular mycorrhizal fungi under a maize cropping system. Plant Soil 270(1):371–382. https://doi.org/10.1007/s11104-004-1829-4
Bloch SE, Clark R, Gottlieb SS, Wood LK, Shah N, Mak SM, Lorigan JG, Johnson J, Davis-Richardson AG, Williams L, McKellar M, Soriano D, Petersen M, Horton A, Smith O, Wu L, Tung E, Broglie R, Tamsir A, Temme K (2020) Biological nitrogen fixation in maize: optimizing nitrogenase expression in a root-associated diazotroph. J Exp Bot 71(15):4591–4603. https://doi.org/10.1093/jxb/eraa176
Calvo-Garrido C, Viñas I, Usall J, Rodríguez-Romera M, Ramos MC, Teixidó N (2014) Survival of the biological control agent Candida sake CPA-1 on grapes under the influence of abiotic factors. J Appl Microbiol 117(3):800–811. https://doi.org/10.1111/jam.12570
Chen Q, Hou LW, Duan WJ, Crous PW, Cai L (2017) Didymellaceae revisited. Stud Mycol 87:105–159. https://doi.org/10.1016/j.simyco.2
Coninck E, Scauflaire J, Gollier M, Liénard C, Foucart G, Manssens G, Munaut F, Legrève A (2020) Trichoderma atroviride as a promising biocontrol agent in seed coating for reducing Fusarium damping-off on maize. J Appl Microbiol 129(3):637–651. https://doi.org/10.1111/jam.14641
Czarnecka M, Żarowska B, Połomska X, Restuccia C, Cirvilleri G (2019) Role of biocontrol yeasts Debaryomyces hansenii and Wickerhamomyces anomalus in plants’ defence mechanisms against Monilinia fructicola in apple fruits. Food Microbiol 83:1–8. https://doi.org/10.1016/j.fm.2019.04.004
de Goes KCGP, da Silva JJ, Lovato GM et al (2017) Talaromyces sayulitensis, Acidiella bohemica and Penicillium citrinum in Brazilian oil shale by-products. Antonie Van Leeuwenhoek 110:1637–1646. https://doi.org/10.1007/s10482-017-0913-8
Delmont TO, Robe P, Cecillon S, Clark IM, Constancias F, Simonet P, Hirsch PR, Vogelet TM (2021) Accessing the soil metagenome for studies of microbial diversity. Appl Environ Microbiol 4:1315–1324. https://doi.org/10.1128/AEM.01526-10
Duniere L, Xu S, Long J, Elekwachi C, Wang Y, Turkington K, Forster R, McAllister TA (2017) Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage. BMC Microbiol 17:50. https://doi.org/10.1186/s12866-017-0947-0
Ekpakpale DO, Kraak B, Meijer M, Ayeni KI, Houbraken J, Ezekiel CN (2021) Fungal diversity and aflatoxins in maize and rice grains and cassava-based flour (Pupuru) from Ondo State. Nigeria J Fungi (basel) 7(8):635. https://doi.org/10.3390/jof7080635
El-Ghwas DE, Elkhateeb WA, Akram M and Daba GM (2014) Yeast as biotechnological tool in food industry. J Pharm Res 5(3):1–6. https://doi.org/10.23880/oajpr-16000243
EUROSTAT (2022) https://ec.europa.eu/eurostat/web/agriculture/data/database. Accessed Feb 2022
Fadiji AE, Ayangbenro AS, Babalola OO (2020) Organic farming enhances the diversity and community structure of endophytic archaea and fungi in maize plant: a shotgun approach. Soil Sci Plant Nutr 20:2587–2599. https://doi.org/10.1007/s42729-020-00324-9
Fadiji AE, Babalola OO (2020) Metagenomics methods for the study of plant-associated microbial communities: a review. J Microbiol Methods 170:105860
FAOSTAT (2022) https://www.fao.org/statistics/en/. Accessed Feb 2022
Gałązka A, Grządziel J (2018) Fungal genetics and functional diversity of microbial communities in the soil under long-term monoculture of maize using different cultivation techniques. Front Microbiol 9:76. https://doi.org/10.3389/fmicb.2018.00076
Genomed (2020) https://www.genomed.pl. Accessed Feb 2020
González-Prieto JM, Rosas-Quijano R, Domínguez A, Ruiz-Herrera J (2014) The UmGcn5 gene encoding histone acetyltransferase from Ustilago maydis is involved in dimorphism and virulence. Fungal Genet Biol 71:86–95. https://doi.org/10.1016/j.fgb.2014.09.002
Guler NS, Pehlivan N, Karaoglu SA, Guzel S, Bozdeveci A (2016) Trichoderma atroviride ID20G inoculation ameliorates drought stress-induced damages by improving antioxidant defence in maize seedlings. Acta Physiol Plant 38:132. https://doi.org/10.1007/s11738-016-2153-3
Henry A, Kosola KR (1999) Root age and phosphorus effects on colonization of Andropogon gerardii by mycorrhizal fungi. Soil Biol Biochem 31:1657–1660. https://doi.org/10.1016/s0038-0717(99)00077-2
Huang L, Kim KT, Yang JY, Song H, Choi G, Jeon J, Cheong K, Ko J, Xu H, Lee YHA (2019) High-Quality Draft genome sequence of Colletotrichum gloeosporioides sensu stricto SMCG1#C, a causal agent of anthracnose on Cunninghamia lanceolata in China. Mol Plant Microbe Interact 32(2):139–141. https://doi.org/10.1094/MPMI-05-18-0144-A
IUSS Working Group WRB (2015) World reference base for soil resources 2014, update 2015. International soil classification system for naming soils and creating legends for soil maps. World Soil Resources Reports No. 106, FAO, Rome, 192 pp
Janisiewicz WJ, Petersen DL, Bors S (1994) Control of storage decay of apples with Sporobolomyces roseus. Plant Dis 78(5):466. https://doi.org/10.1094/PD-78-0466
Jastrzębska M, Wachowska U, Kostrzewska MK (2020) Pathogenic and non-pathogenic fungal communities in wheat grain as influenced by recycled phosphorus fertilizers: a case study. Agriculture 10(6):239. https://doi.org/10.3390/agriculture10060239
Johnson D, Vandenkoornhuyse PJ, Leake JR, Gilbert L, Booth RE, Grime JP, Peter J, Young W, Read DJ (2003) Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosms. New Phytol 161:503–515. https://doi.org/10.1046/j.1469-8137.2003.00938.x
Keirnan CE, Tan YP, Laurence MH, Mertin AA, Edward CY, Liew ECY, Summerell BA, Shivas RG (2021) Cryptic diversity found in Didymellaceae from Australian native legumes. MycoKeys 78:1–20. https://doi.org/10.3897/mycokeys.78.60063
Köhl J, Kolnaar R, Ravensberg WJ (2019) Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front Plant Sci 10:845. https://doi.org/10.3389/fpls.2019.00845
Kong X, Han Z, Tai X, Jin D, Ai S, Zheng X, Bai Z (2020) Maize (Zea mays L. sp.) varieties significantly influence bacterial and fungal community in bulk soil, rhizosphere soil and phyllosphere. FEMS Microbiol Ecol 96(3):020. https://doi.org/10.1093/femsec/fiaa020
Kuchar L, Szwejkowski Z, Rychcik B, Dragańska E, Cymes I (2021) Regional climate changes in north-eastern Poland and their impact on maize crops. Acta Sci Pol Technol Aliment 20(1):3–18. https://doi.org/10.15576/ASP.FC/2021.20.1.3
Kusuma A, Widiastuti A, Priyatmojo A (2017) Induction of resistance using Trichoderma spp. and Penicillium sp. against banded leaf and sheath blight (BLSBb) caused by Rhizoctonia solani in maize. In: Isnansetyo A., Nuringtyas T.R. (eds.), Proceeding of the 1st International Conference on Tropical Agriculture. 0.1007/978–3–319–60363–6_1
Lević J, Petrović T, Stanković S, Ivanović D (2013) The incidence of Pyrenochaeta terrestris in root of different plant species in Serbia. Zb. Matice Srp. za Prir. Nauk. / Matica Srp. J Nat Sci 125:21–30. https://doi.org/10.2298/ZMSPN1325021L
Li B, Peng H, Tian S (2016) Attachment capability of antagonistic yeast Rhodotorula glutinis to Botrytis cinerea contributes to biocontrol efficacy. Front Microbiol 7:601. https://doi.org/10.3389/fmicb.2016.00601
Liu J, Yao Q, Li Y, Zhang W, Mi G, Chen X, Yu Z, Wang G (2019) Continuous cropping of soybean alters the bulk and rhizospheric soil fungal communities in a Mollisol of Northeast PR China. Land Degrad Dev 30(14):1725–1738. https://doi.org/10.1002/ldr.3378
Liu XZ, Wang QM, Göker M, Groenewald M, Kachalkin AV, Lumbsch HT, Millanes AM, Wedin M, Yurkov AM, Boekhout T, Bai FY (2015) Towards an integrated phylogenetic classification of the Tremellomycetes. Stud Mycol 81:85–147. https://doi.org/10.1016/j.simyco.2015.12.001
Mahmood T, Azam F, Malik KA (1986) Decomposition and humification of plant residues by some soil fungi. Biotech Lett 7(3):207–212
Malicka M, Magurno F, Posta K, Chmura D, Piotrowska-Seget Z (2021) Differences in the effects of single and mixed species of AMF on the growth and oxidative stress defense in Lolium perenne exposed to hydrocarbons. Ecotoxicol Environ Saf 217(1):112252. https://doi.org/10.1016/j.ecoenv.2021.112252
Mao L, Liu Y, Zhang J, Okerblad J, Chen S, Johnson NC (2021) Soil biota suppress maize growth and influence root traits under continuous monoculture. Plant Soil 461:441–455. https://doi.org/10.1007/s11104-021-04848-6
Marin-Felix Y, Groenewald JZ, Cai L et al (2017) Genera of phytopathogenic fungi: GOPHY 1. Stud Mycol 86:199–216. https://doi.org/10.1016/j.simyco.2017.04.002
Mašínová T, Bahnmann BD, Větrovský T, Tomšovský M, Merunková K, Baldrian P (2017) Drivers of yeast community composition in the litter and soil of a temperate forest. FEMS Microbiol Ecol 93(2):fiw223. https://doi.org/10.1093/femsec/fiw223
Meier U (1997) Growth stages of mono- and dicotyledonous plants: BBCH-Monograph. Blackwell Wissenschafts-Verlag, Berlin, p 622
Moreno MV, Casas C, Biganzoli F, Manso L, Silvestro LB, Moreira E, Stenglein SA (2021) Cultivable soil fungi community response to agricultural management and tillage system on temperate soil. Saudi Soc Agricul Sci 20(4):217–226. https://doi.org/10.1016/j.jssas.2021.01.008
Mwakilili AD, Mwaikono KS, Herrera SL et al (2021) Long-term maize-Desmodium intercropping shifts structure and composition of soil microbiome with stronger impact on fungal communities. Plant Soil 467:437–450 (10.1007)
Naef A, Défago G (2006) Population structure of plant-pathogenic Fusarium species in overwintered stalk residues from Bt-transformed and non-transformed maize crops. Eur J Plant Pathol 116(2):129–143. https://doi.org/10.1007/s10658-006-9048-x
Nandya SK, Srivastavab RK (2018) A review on sustainable yeast biotechnological processes and applications. Microbiol Res 207:83–90. https://doi.org/10.1016/j.micres.2017.11.013
Narayanasamy P (2013) Mechanisms of action of fungal biological control agents. In: Biological management of diseases of crops. PIBC (Progress in Biological Control) 15:99–200
Ozimek E, Hanaka A (2021) Mortierella Species as the Plant Growth-Promoting Fungi Present in the Agricultural Soils. Agriculture 11(1):7. https://doi.org/10.3390/agriculture11010007
Pathi KM, Rink P, Budhagatapalli N, Betz R, Saado I, Hiekel S, Becker M, Djamei A, Kumlehn J (2020) Engineering smut resistance in maize by site-directed mutagenesis of LIPOXYGENASE 3. Front Plant Sci 11. https://doi.org/10.3389/fpls.2020.543895
Pechanova O, Pechan T (2015) Maize-pathogen interactions: an ongoing combat from a proteomics perspective. Int J Mol Sci 16(12):28429–28448. https://doi.org/10.3390/ijms161226106
Pinzari F, Maggi O, Lunghini D, Di Lonardo DP, Persiani AM (2017) A simple method for measuring fungal metabolic quotient and comparing carbon use efficiency of different isolates: Application to Mediterranean leaf litter fungi. Plant Biosyst 151(2):371–376. https://doi.org/10.1080/11263504.2017.1284166
Roumet C, Birouste M, Picon-Cochard C, Ghestem M, Osman N, Vrignon-Brenas S, Cao KF, Stokes A (2016) Root structure-function relationships in 74 species: Evidence of a root economics spectrum related to carbon economy. New Phytol 210(3):815–826. https://doi.org/10.1111/nph.13828
Rychcik B, Adamiak J, Wójciak H (2006) Dynamics of the soil organic matter in crop rotation and long-term monoculture. Plant Soil Environ 52(Special Issue):15–20
Sasvári Z, Hornok L, Posta K (2011) The community structure of arbuscular mycorrhizal fungi in roots of maize grown in a 50-year monoculture. Biol Fertil Soils 47(2):167–176. https://doi.org/10.1007/s00374-010-0519-z
Saveetha K, Nimalka W, Geetha SR, Greg TR, Patterson G, Shimaila A, George L (2021) Disentangling the association of corn root mycobiome with plant productivity and the importance of soil physicochemical balance in shaping their relationship. Front Sustain Food Syst 5(17). https://doi.org/10.3389/fsufs.2021.617332
Schisler DA, Yoshioka M, Vaughan MM, Dunlap CA, Rooney AP (2019) Nonviable biomass of biocontrol agent Papiliotrema flavescens OH 182.9 3C enhances growth of Fusarium graminearum and counteracts viable biomass reduction of Fusarium head blight. Biol Control 128:48–55. https://doi.org/10.1016/j.biocontrol.2018.09.006
Smith SE, Smith FA (2012) Fresh perspectives on the roles of arbuscular mycorrhizal fungi in plant nutrition and growth. Mycologia 104(1):1–13 (http://www.jstor.org/stable/23055298)
Sobowale AA, Odebode AC, Cardwell KF, Bandyopadhyay R, Jonathan SG (2010) Antagonistic potential of Trichoderma longibrachiatum and T. hamatum resident on maize (Zea mays) plant against Fusarium verticillioides (Nirenberg) isolated from rotting maize stem. Arch Phytopathol 43(8):744–753. https://doi.org/10.1080/03235400802175904
Standen JH (1945) Nigrospora oryzae (B. and Br.) Fetch on Maize. Phytopathology 35(7):553–564
StatSoft (2019) TIBCO Software Inc., 2019. Statistica (data analysis software system), version 13. https://www.statsoft.pl/
Strom N, Hu W, Haarith D, Chen S, Bushley K (2020) Interactions between soil properties, fungal communities, the soybean cyst nematode, and crop yield under continuous corn and soybean monoculture. Appl Soil Ecol 147:103388. https://doi.org/10.1016/j.apsoil.2019.103388
Sugawara F, Strobel G, Strange RN, Siedow JN, Van Duyne GD, Clardy J (1987) Phytotoxins from the pathogenic fungi Drechslera maydis and Drechslera sorghicola. Proc Natl Acad Sci 84(10):3081–3085
Tanwar A, Aggarwal A, Yadav A, Parkash V (2013) Screening and selection of efficient host and sugarcane bagasse as substrate for mass multiplication of Funneliformis mosseae. Biol Agric Hortic 29(2):107–117. https://doi.org/10.1080/01448765.2013.771955
Tyagi S, Mandal SK, Kumar R, Kumar S (2018) Effect of different herbicides on soil microbial population dynamics in rabi maize (Zea mays L.). Int J Curr Microbiol App Sci Special Issue 7:3751–3758
van Capelle C, Meyer-Wolfarth F, Meiners T, Schrader (2021) Lumbricus terrestris regulating the ecosystem service/disservice balance in maize (Zea mays) cultivation. Plant Soil 462:459–475. https://doi.org/10.1007/s11104-021-04882-4
van Reeuwijk LP (2002) Procedures for soil analysis, sixth ed. Technical Papers Wageningen, the Netherlands, ISRIC - World Soil Information, 9:120
Wachowska U, Stuper-Szablewska K, Perkowski J (2020) Yeasts isolated from wheat grain can suppress Fusarium head blight and decrease trichothecene concentrations in bread wheat and durum wheat grain. Pol J Environ Stud 29(6):4345–4360. https://doi.org/10.15244/pjoes/118427
Wang C, White PJ, LiC C (2017) Colonization and community structure of arbuscular mycorrhizal fungi in maize roots at different depths in the soil profile respond differently to phosphorus inputs on a long-term experimental site. Mycorrhiza 27(4):369–381. https://doi.org/10.1007/s00572-016-0757-5
Wang L, Wang X, Gao F, Lv C, Li L, Han T, Chen F (2021) AMF Inoculation can enhance yield of transgenic Bt maize and its control efficiency against Mythimna separata especially under elevated CO2. Front Plant Sci 12. https://doi.org/10.3389/fpls.2021.655060
Woźniak A (2019) Effect of crop rotation and cereal monoculture on the yield and quality of winter wheat grain and on crop infestation with weeds and soil properties. Int J Plant Prod 13:177–182. https://doi.org/10.1007/s42106-019-00044-w
Acknowledgements
Profound thanks go to Professor Witold Niewiadomski and Professor Kazimiera Zawiślak who established a field experiment investigating crop rotation and monoculture in 1967. We would also like to thank the staff of the Experimental Station in Bałcyny for conducting the field experiment on maize cultivation for more than 50 years.
Funding
Project financially supported by the Minister of Education and Science under the program entitled “Regional Initiative of Excellence” for the years 2019–2023, Project No. 010/RID/2018/19, amount of funding 12 000 000 PLN.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Urszula Wachowska. The first draft of the manuscript was written by Urszula Wachowska and Bogumił Rychcik. All authors commented on previous versions of the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Stéphane Compant.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wachowska, U., Rychcik, B. Plants control the structure of mycorrhizal and pathogenic fungal communities in soil in a 50-year maize monoculture experiment. Plant Soil 484, 133–153 (2023). https://doi.org/10.1007/s11104-022-05779-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-022-05779-6