Abstract
Background
Plant diseases significantly affect the crop, so their identification is very important. Correct identification of these diseases is crucial for establishing a good disease control strategy to avoid time and financial losses. In general, machines can greatly reduce the possibility of human error. In particular, computer vision techniques developed through deep learning have paved a way to detect and diagnose these plant diseases on the leaf.
Methods
In this work, the model AFD-Net was developed to detect and identify various leaf diseases in apple trees. The dataset is from Kaggle 2020 and 2021 and was financially supported by the Cornell Initiative for Digital Agriculture. An AFD-Net was proposed for leaf disease classification in apple trees and the results of the efficiency of the model are compared with other state-of-the-art deep learning approaches.
Results
The results of the experiments in the validation dataset show that the proposed AFD-Net model achieves the highest values of 98.7% accuracy for Plant Pathology 2020 and 92.6% for Plant Pathology 2021 compared to other deep learning models in the original and extended datasets.
Discussion
The results also indicate the efficiency of the proposed model in identifying leaf diseases on apple trees for major and minor classes, i.e., for multiple classification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaf diseases of apples refer to the common diseases found on apple leaves, namely scab, rust, and powdery mildew, among others (Thapa et al. 2020). Apple scab, caused by a fungal pathogen, is one of the most economically important fungal diseases of apple in the world (Agarwal et al. 2019; Barbedo 2018). The symptoms of apple scab are clearly visible fungal structures on the surface of the leaf. Rust disease also causes severe losses when environmental conditions are favorable for disease growth. For example, in a plant affected by Rust (Moinina et al. 2019), small yellow spots appear on the leaf surface.
Farmers spend a lot of money on disease control and inadequate technical support, but the results always lead to poor disease control (Huber and Jones 2013; Husson et al. 2021). Foliar diseases spread rapidly and can destroy large portions of the yield in a very short time. In some cases, these diseases destroy the entire crop if the disease is not controlled quickly and accurately. Foliar diseases are a challenge to crop production in most countries. They reduce crop yields, fruit quality, and nutritional value, resulting in lower returns for the farmers (Moinina et al. 2019).
Machine learning models learn, recognize patterns, and make decisions with minimal human intervention (Bhateja et al. 2018; Raj et al. 2011). Ideally, machines increase accuracy and efficiency and eliminate the possibility of human error (Zhong and Zhao 2020). The use of AI in agriculture helps farmers gain insights into their crops and use the data to increase their overall production. Various computer vision techniques can be applied to gain the desired insights (Militante et al. 2019; Raschka 2018; Chaki et al. 2019).
Recent advances in computer vision enabled by deep leaning have paved the way for more accurate disease diagnosis. Using large public datasets of diseased and healthy plants and leaves, a CNN can be trained to identify various leaf diseases (Agarwal et al. 2019; Moinina et al. 2018; Tharwat et al. 2016). With the increasing availability of smartphones, the approach of training deep-learning models on a large scale has emerged as a clear way to diagnose crops on a large scale (Zhong and Zhao 2020). Every year, farmers worldwide are affected by foliar diseases. Our research could contribute greatly to the automation of disease detection worldwide and potentially help millions of people (Thapa et al. 2020).
Major crops today are plagued by a variety of diseases. Diseases in crops can occur in various parts of the plant, such as the roots, leaves, and stem, although the leaves are the most typical site for disease detection. It is difficult to detect and diagnose diseases because leaves have a variety of sizes, shapes, and colors. The article deals with various aspects of diseases and classifies them based on the characteristics of the condition of the leaves. It is important to identify the cause at the root, which is beneficial and time-saving for both the agricultural sector and farmers. So far, to our knowledge, there are some works on leaf disease detection in apples, and these datasets have fewer classes (Agarwal et al. 2019; Zhong and Zhao 2020). Only one work has looked at the 2020 plant pathology dataset, but it has the limitation of lacking augmentation techniques. Also, there was no cross-validation to determine the out-of-fold predictions and the scores of the respective folds (Thapa et al. 2020). This could lead to an overfitting problem in some cases because the model does not have adequate validation for evaluation.
Our approach is to first classify a given image from a test dataset to identify the conditions of the plants (i.e., diseased or healthy). Moreover, the dataset is preprocessed and extensions are made. After that, different diseases can be identified and sorted out using the proposed model. The results obtained by applying the proposed model can lead to multiple diseases on a plant (i.e., more than one). The main contribution of this paper is as follows:
-
1.
This paper discusses the current state-of-the-art of machine learning and deep learning applications in disease identification and classification. A novel AFD-Net model is proposed to automate the detection and multiple classification of foliar diseases in apples using the Hybrid EfficientNet model.
-
2.
AFD-Net model connect lambda layers B3 and B4 layers of the network followed by a dropout layer to prevent overfitting of the model. The use of a dense layer with 4 units and a softmax activation function as the last layer completes the model architecture.
-
3.
The proposed AFD-Net model achieves 98.7% and 92.6% accuracy for apple foliar disease in the plant pathology 2020 and plant pathology 2021 datasets, respectively. The achieved performance outperforms other state-of-the-art deep learning and transfer learning models.
In addition, Literature Review section provides a detailed studies on leaf disease detection and classification in apples using machine learning and deep learning methods. In Apple Foliar Disease Neural Network section, the proposed methodology “AFD-Net” and the evaluation parameters are explained in detail. Moreover, the description of the dataset and its preprocessing steps are discussed in Dataset Description and Pre-processing. In Experimental Evaluation, the design of the implementation and the analysis of the results are described and discussed. Finally, conclusions are drawn and future work is described in Discussion and Conclusion.
Literature review
In recent years, ML and DL are widely used to detect plant diseases, which helps farmers identify the right foliar disease and apply the appropriate treatments (Mahlein 2016). Digital images are widely used in computer vision to identify the diseases for further classification based on their symptoms (Barbedo 2014; Dai et al. 2019; Wöhner and Emeriewen 2019; Sladojevic et al. 2016). However, it is challenging to accurately identify disease from leaves due in part to the resolution, background light, and shadows of the leaves, among other (Jadhav et al. 2020). Machine learning (ML) and deep learning (DL) approaches are well suited for processing image data, especially in agriculture, and can be used to detect and classify plant diseases from the collected images, i.e., photos of leaves (Amara et al. 2017).
Agarwal et al. proposed a model consisting of 3 maximal pooling layers followed by two densely connected layers. After testing with different numbers of convolutional layers from 2 to 6, it was found that 3 layers provide the best accuracy (Agarwal et al. 2019). The proposed model achieves a very impressive accuracy, i.e., 96%. The database used for the developed framework consists of nearly 50,000 images of 171 diseases, including 21 plant species. The original samples were divided into smaller images containing individual lesions or localized symptom regions. This was done to increase the size of the dataset and to test how the CNN would perform with more localized information.
Instead of taking pictures in the natural condition, Zhong and Zhao took pictures with a solid background (Zhong and Zhao 2020). Images of all symptoms were resized to 128 \(\times\) 128. The dataset was split 8:2 for the training and test dataset by randomly selecting images from the dataset. After duplication, the dataset contained 2,462 images, with 85% of the images used for training and 15% for validation. The accuracy of this method for the test dataset was 93.71%.
Militante et al. proposed a model, i.e., a combination of a convolutional layer, an activation layer, a pooling layer, and a fully connected layer (Militante et al. 2019). The images used in this study were in color and were reduced to 96 by 96 for further processing. An accuracy of 96.5% was achieved with 75 epochs while the model was well trained. A maximum accuracy of 100% was achieved when random images of plant varieties and diseases were tested.
Sladojevic et al. presented a dataset of 79,265 images (Sladojevic et al. 2016). Traditional augmentation methods and generative adversarial networks are used for image augmentation. Moreover, a 2-stage NN architecture was proposed for classification and a test accuracy of 93.67% was achieved with the trained model. The DCNN model (Chao et al. 2020) was presented for detecting the leaf disease of apple tree by combining DenseNet and Xception. The results show that the developed model achieved 98% accuracy.
Yu et al. designed two subnetworks; the first is used for segmentation to identify features, and the second model is used for classification. In the experiments, the proposed model provided an accuracy of 89.4% (Yu et al. 2020). The use of AI in agriculture helps farmers gain insights about their crops and use this data to increase their production wisely. Using the proposed methods, Thapa et al. captured 3,650 of high-resolution images of several apple leaf diseases and annotated the dataset with the help of an expert in the field of pathology to confirm the annotations for the images, which were difficult to distinguish based on symptoms (Thapa et al. 2020). The overall test accuracy achieved by a ResNet50 network pre-trained on ImageNet was 97%.
Raschka et al. proposed a CNN model and achieved 97.62% accuracy in identifying four different types of apple leaf blight, detecting infected parts on the leaf, and classifying between healthy and infected fruits, e.g., apples (Raschka 2018). However, there are very few studies dealing with apple foliar disease and most of them are limited to a specific type of disease, either biotic or abiotic.
Apple foliar disease neural network
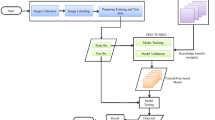
In this section, a framework called Apple Foliar Disease Neural Network (AFD-Net) is presented in this paper. Basically, our work is based on the transfer learning approach, where the first and most important step is to collect the dataset (Thapa et al. 2020). The second step is to project and clean the database using image processing steps (Guo et al. 2008) to find outliers and class imbalances. This was followed by an exploratory data analysis of the dataset with all graphs and class distribution of the foliar diseases. Extensions such as rotation, transformation, and flips were applied to increase the diversity/learning capability of the model (Yun et al. 2019; Zhang et al. 2017). After preprocessing all the data, the data were fed into the training pipeline with fivefold cross-validation, properly validating the training data. These iterations are repeated continuously until we find a stable cross-validation value for our training data that matches the test data.
As shown in Fig. 1, the model takes the image dataset as input. In the next step, the image data is preprocessed and further augmentation and transformations generate the final image data to be processed by the model. The model is then fed with this processed image data along with the metadata about the input image dataset. Finally, this collected input is processed by the AFD-Net model, which consists of neural layers with lambda layers B3 and B4 combined, followed by a dropout layer to prevent overfitting of the model. The use of a dense layer with 4 units and a softmax activation function as the last layer completes the model architecture.
To improve the performance of the model, several strategies can be considered, e.g., hyper-parameter tuning approaches, changing the loss function such as bi-tempered loss for noisy labels, changing the learning rate, and freezing/unfreezing the model layers. Different ensemble approaches (weighted average, normal average, rank aggregate) were also considered to increase the prediction accuracy. In the designed model, the weighted average strategy was chosen as the model for the designed approach. The flowchart for the above methodology is shown in Fig. 1.
Efficient net distribution
The performance of the models used in the ImageNet dataset has increased since 2012 (Tan and Le 2019) as they have become more complex, but many of them are not effective in terms of computational load. The EfficientNet model (Tan and Le 2019), which is one of the best models since it achieves 84.4% accuracy on the ImageNet classification problem with 66 M parameters, can be considered as a group of CNN models. It consists of 8 models between B0 and B7, and as the number of models increases, the number of computed parameters does not increase significantly while the accuracy increases noticeably (Tan and Le 2019). We used only B3 and B4 to find a middle ground, as they performed the best under various experiments and hardware constraints. Unlike other CNN models, EfficientNet uses a new activation function called Swish instead of the Rectifier Linear Unit (ReLU) activation function (Tan and Le 2019). From the experiments, simply substituting ReLU units with Swish units improves the classification accuracy in ImageNet by 0.6% for Inception-ResNet-V2; it outperforms ReLU in many deep neural networks, as shown in Fig. 2.
Architecture of the proposed AFD-Net
In the proposed AFD-Net model, EfficientNet B3 and B4 are attached along with the lambda layer, which is capable of running on few parameters to obtain great results with ensemble of the probabilities from both the architectures. At the end Dropout layers were added to the above functional layers of the model, and ended with softmax output function. In total, there were 28.2 M trainable parameters in the experiments. The AFD-Net achieve both high accuracy and efficiency over pre-existing CNN based models, this reducing parameter size and achieving the accuracy. The architecture of developed AFD-Net can be seen in Fig. 3.
In this proposed model of lambda layers, we connected two functional layers of efficient nets B3 and B4, followed by Dropout layer to prevent overfitting of the model, and at the end we added a Dense layer of 4 units together with a softmax activation function for the prediction part. The developed AFD-Net is then described in Algorithm 1.

The algorithm takes as input the set of images as t-f records with labels. These images are passed through the augmentation module to extract a variety of augmented images that are fed into the training module of the AFD-Net. The augmentations are of different types and range from flipping to cropping. The resulting augmented images and their associated labels are then converted into one-hot encoding vectors with labels to incorporate the categorical crossentropy loss (lines 1 to 2). After these preprocessing steps, the data are split into a training set and a validation/test set using K-fold cross-validation with 5 folds stratified by labels (line 4). In lines 5 to 10, each tuple of the training dataset is fed into the AFD-Net model, setting the learning rate and other hyperparameters of the model (line 6). In the AFD-Net model, EfficientNet B3 and B4 are attached along with the lambda layer, which can work with few parameters to produce great results with an ensemble of probabilities from both architectures (line 7). In the end, Dropout layers were added to the above functional layers of the model and finished with the softmax output function to generate probabilities for each label/class. Finally, the validation/test tuples are used to make the predictions about the model and store the results in a data frame (lines 8 to 9). Finally, the trained model is saved to disk after all epochs are completed (line 11).
Evaluation metrics
The efficiency of the proposed model is evaluated with fivefold cross-validation using stratified splits (Ayaz et al. 2021). The dataset was split into two parts, i.e., a training set and a validation set. The validation part is used to test the model. The performance metrics selected in this study are commonly used to measure model efficiency and performance, including accuracy, confusion matrix, specificity, sensitivity, precision, F1-score, and Matthews correlation coefficient (MCC). The determined parameters of these metrics are based on the rates of true positive (x), true negative (y), false positive (z), and false negative (w), as shown in Table 1.
Transfer learning algorithm for comparison
Inception-v3
Inception-v3 (Szegedy et al. 2016) is a design of a convolutional neural network from the Inception family that includes label smoothing, factorized 7 \(\times\) 7 convolutions, and the inclusion of an auxiliary classifier to propagate label information further down the network. Inception-v3 proposed a method for regularizing the classification layer by computing the marginalized impact of label dropout during training. The smoothing of the label prevents overfitting of the model. The equation is then shown in Eq. (1)
where \(\epsilon\) is a hyper parameter and set as 0.1 and N is the number of classes and set it as 4.
ResNet50 and ResNet101
When training deep networks, there comes a moment when accuracy reaches a saturation point and then rapidly degrades. This shows that not all neural network designs can be optimized equally well. To solve this problem, ResNet (He et al. 2016) employs a method known as “residual mapping”. The residual network allows these layers to explicitly match a residual mapping, rather than trusting that all pairs of stacked layers would match a desired underlying mapping. The building block of a residual network is shown below in Fig. 4.
In addition, the feedforward neural networks with shortcut connections can be then formalized as:
VGG-16
Convolutional layers one and two have a kernel of 64 features with 3 \(\times\) 3 filter size. Convolutional layers three and four have a kernel of 124 features with 3 \(\times\) 3 filter size. Two layers are followed by a 2 max-pool layer, so 56 \(\times\) 56 \(\times\) 128 will be the reduced output of the results. For the fifth, sixth and seventh layers, 256 feature maps with a size of 3 \(\times\) 3 are used. Followed by 2 max-pool layers, 512 filters will be used for the eighth through thirteen convolutional layers with kernel size 3 \(\times\) 3. Followed by one max-pool layer. There are fully connected hidden layers with 4096 units, followed by a softmax output layer with 1, 000 units (He et al. 2016).
Dataset description and pre-processing
The dataset (Thapa et al. 2020) used in this paper was taken from the Plant Pathology 2020-FGVC7 Kaggle competition, which was financially supported by the Cornell Initiative for Digital Agriculture (CIDA). The dataset consists of 1, 821 portrait and landscape images. The image size is either 2,048 \(\times\) 1,368 pixels or 1,368 \(\times\) 2,048. The second dataset is that of the Plant Pathology 2021-FGVC8 competition with a pilot dataset of 3, 651 RGB images of leaf diseases in apples. For Plant Pathology 2021-GVC81, the number of leaf disease images was significantly increased and additional disease categories were added. The dataset contains 18,633 of high-quality RGB images of leaf diseases in apples, including a large, expert-annotated disease dataset. This dataset reflects real field scenarios by representing in homogeneous backgrounds of leaf images taken at different stages of ripening and at different times of day with different camera settings. We generated images of dimensions 512 \(\times\) 512 with two string and int byte structures for images, names, and targets. The generated images were set to 100% quality. Along with the images, the partitioning was layered into 15 folds to maintain the proper class balance throughout the dataset.
Target distribution
We have 4 categories of foliage in the Plant Pathology 2020 data set, including “healthy”, “rust”, “scab”, and “multiple Diseases”. “rust” is the prominent disease, followed by “scab”, “healthy”, and the lowest number of “multiple Diseases i.e., C1 to C4. It was noted that the data were unbalanced with respect to the Multiple Diseases class (see Fig. 5). In addition, the 2021 plant pathology dataset is categorized into 8 target classes. All target classes and their number distributions are shown in Fig. 6.
Channel distribution findings
Green is the most prominent color in the leaf record, which makes sense since the leaves are colored green. There was a lot of variance in the dataset for the red and blue channels in both cases. The variance occurred in the infected leaves that were affected by scab, rust, or multiple diseases. The channel values appear to have an approximately normal distribution centered around 105. The green portions of the image have very low blue values, but in contrast, the brown portions have high blue values. This indicates that the green (healthy) parts of the image have low blue values, while the unhealthy parts tend to have high blue values. An unhealthy leaf with its RGB values is shown in Fig. 7.
Data splitting
In this study, the original datasets of apple leaf diseases “Plant Pathology 2020” and “Plant Pathology 2021” are used. The datasets are randomly divided into training and validation sets, i.e., 80% and 20%, respectively. The training and validation sets were used only for fitting the model and calculating the validation metrics, which can be seen in Table 2. The original apple foliar disease dataset “Plant Pathology 2020” and “Plant Pathology 2021” are used in this study. The datasets are randomly divided into training and validation sets i.e., 80% and 20%, respectively. Training and validation sets were only used for fitting of the model and calculating validation metrics respectively see Table 2.
For cross-validation, we used 5 folds split across 15 training datasets, each consisting of 120 targets (approx.)—healthy, rust, scab, and multiple diseases, respectively, for the original dataset. The training and validation datasets were only used to fit the model and calculate out-of-fold predictions, respectively.
Cutmix, mixup and basic augmentations
To help the model learn all the outliers and reveal the state of the leaves, it must be trained with different augmentation images to increase its robustness. This will make the model more flexible to the newly injected and eccentric data. Different types of augmentations can be used for different problems. For the designed model, we used the basic augmentations provided by the Keras module—random (crop, hue, saturation, brightness and contrast) and shear transform, as shown in Fig. 8.
The cutMix (Yun et al. 2019) and mixup (Zhang et al. 2017) are the two augmentation techniques we implemented in the dataset. In the cutmix algorithm, part of the image is appended/attached to the other image to improve the localization of the model. Instead of simply cutting out pixels, as is the case with cutout or dropout, we replace the cutout regions with a patch from another image. The ground truth labels are blended in proportion to the pixel count of the composite image. By asking the model to identify the object from a partial view, the additional patches can improve localization. In mixup expansion, two samples are mixed together by linear interpolation of their images and labels. Mixup samples suffer from unrealistic output and label ambiguity, and thus cannot perform well in tasks such as image localization and object recognition. Mixup alleviates this problem by mixing different features together, preventing a network from having too much confidence in the association between features and labels. Dropout/Cutout augmentation is a type of regional dropout strategy in which a random patch from an image is zeroed out (replaced with black pixels). Cutout samples suffer from reduction in information and regularization ability.
Experimental evaluation
In this section, we discuss the detailed implementation of the proposed AFD-Net model. It was implemented using Google Colab and trained with TPUs. The model was tested with several k-folds, of which the 5-folds are the most efficient in terms of metrics. The custom learning scheduler was also optimized based on ramp-up epochs and decay value. Initially, we started with the categorical cross-entropy as the loss function with a label smoothing of 0.05 and then witched to the bi-tempered logistic loss to compare the model performance.
Network training
Before training the network on the leaf dataset, we used pre-trained weights from ImageNet (Tan and Le 2019) and noisy-student. The dataset with image size 512 × 512 was obtained from the training pipeline created for the purpose of pre-fetching tf-records with features such as caching and image decoding. The network was trained with a custom learning rate scheduler applied to the optimizer “adam”. We used two loss functions such as “categorical cross-entropy loss” and “bi-tempered logistic loss” as benchmarks. During hyper-tuning of the parameters, some of the tests were performed with different ratios and k-fold split validation. Finally, the model for the cv scheme was trained on 80–20 splits of the training data for 40 epochs and then trained on fivefold cv with 15-stratified splits over classes for 100 epochs per fold. During training, the label smoothing parameter in the loss functions helped the model stabilize and reduce predictions.
Performance comparison
Table 3 shows the various models tried with the image data. Comparing the different models from Table 3, it can be seen that using cutmixup can lead to a significant drop in accuracy compared to dropout augmentation, i.e., 88.4% from 96.7%. In addition, using efficient nets B3 and B4 layers separately leads to lower accuracy compared to combining these two i.e., 91.8% and 93.2%, respectively. It can also be inferred that the weights of noisy-student were better at extracting features of the dataset and provided a good accuracy of 98.7%, as can be seen in Table 3.
Parameters were tuned for the training phase of the proposed models and their variations. The stack size and seed were kept the same for all experiments to achieve consistent results. The difference is in the hyperparameters, noisy student and ImageNet are the weights provided by Efficient net. The accuracy results listed below for the tuned models are considered the average over all 5-folds.
Although existing deep learning networks such as VGG, Inception, and ResNet can be used for leaf disease classification, these models have limitations in improving discriminatory power because they do not account for the mechanism of spatial attention to extract discriminating features between diseased and non-diseased areas. The accuracy of the AFD-Net model (98.7%) outperforms the existing TL models: ResNet 50(95.2%), ResNet (101 96.3%), VGG16 (96.7%), and Inception (95.6%). Therefore, the proposed “AFD-Net-Noisy Student” outperforms other state-of-the-art transfer learning approaches, which can be seen in Table 4.
In addition, the “Plant Pathology 2021” dataset is used to analyze the performance of the AFD-Net model. From Table 5, the accuracy of the proposed model is 92.6%, outperforming the other TL models and the three winners of the competition, i.e., for 1st place 88.3%, 2nd place 87.98%, and 3rd place 87.56%.
Comparison of proposed AFD-Net with existing approaches
In the literature review, we saw that some authors have worked on apple foliar disease detection. Most authors have worked with different data sets and some with small data sets. To our knowledge, only one article has worked with a plant pathology dataset and achieved an accuracy of 97%. Also, we compared our proposed model with existing models (Thapa et al. 2020; Agarwal et al. 2019; Zhong and Zhao 2020; Yu et al. 2020) for apple foliar disease detection and it is observed that presented AFD-Net achieved an accuracy of 98.7%, which can be seen in Table 6.
Inferential statistical analysis
Inferential statistics were performed on “AFD-Net: noisy-student (cross-validation)”. To evaluate the performance of the designed model, we decided to take the out-of-fold predictions from each fold and compute the previously mentioned metrics. Training of the model was started with a custom learning rate scheduler to maximize validation accuracy and minimize loss as the model converged. The model was consistent after the warm-up epochs and did not deviate significantly from detecting underfitting or overfitting. Although there was a minority in the fourth class, i.e., “multiple diseases”, the model yielded an accuracy of 0.78. From the confusion matrix, it is clear that the model had difficulty with the “multiple diseases” class due to the imbalance between the classes, which is why the recall value for this class was 0.55, as shown in Fig. 9. The curve for accuracy versus recall is also shown in Fig. 10. The precision-recall curve justifies the model’s ability to correctly classify the data into the correct classes. Also, a balanced ROC curve (precision-recall curve) justifies the balance of classes.
Figures 11 and 12 show the training results of the model using fivefold splits. Of the 5 folds, the best fold was considered. For further analysis, we plotted the ROC curve for our proposed AFD-Net for different classes and it was found that the accuracy is around 99% for all classes except for multiple classes as shown in Fig. 13.
In addition, we have calculated a quantitative analysis of the parameters of the proposed model. From the Table 7, we can infer the evaluation metrics. Sensitivity is another name for Recall. It is a measure of the proportion of actual positive cases that were predicted to be positive. In our case, the sensitivity is 1, which means that the proportion of true positives is higher than the proportion of false negatives. Similarly, for specificity, a higher value leads to a higher proportion of true negative cases and a lower rate of false positives. For the measure of accuracy, the values for each class with the lowest score are unique for the “multiple diseases” classification. For the proposed AFD-Net model with LR 5e-06 obtains the precision and recall value close to 1, except for multiple diseases and also the F1 score compared to other loss functions.
Besides, the performance of all the models is compared with that of the proposed model AFD-Net and from the results of the Table 8, the proposed model performs better than the other approaches. For our proposed model, the values of sensitivity (Se), specificity (Sp), precision (P), F1 score (F1), accuracy (A) and MCC are 0.99, 0.97, 0.89, 98.7 and 0.94, respectively. These values proved to be better than those of other models. For better illustration, we have also plotted a graph for all the models. It can be clearly seen that the performance of our proposed model is better than the other models for both loss functions, i.e., LR 5e-06 and LR 5e-03, that can be observed from Figs. 14 and 15.
Discussion and conclusion
The world around us relies heavily on the agricultural sector to provide food. Early detection of plant diseases is critical to the industry. In this article, AFDNet model is proposed to identify leaf disease in apple trees. The proposed model is applied to two data sets: Plant Pathology 2020 and Plant Pathology 2021. The model clubs the lambda layers of the neural net model with B3 andB4 layers which significantly enhance the performance of the model. In general, model’s performance can be expressed as: (1) The proposed AFD-Net model achieves an accuracy of 98.7%, which is higher than that of other transfer learning models (B3, B4, Inception V3, VGG16, ResNet50, and 101). (2) The performance of the proposed model also outperforms the other deep learning-based models for both datasets (see Table 3). (3) The obtained results show he efficiency of the proposed model in identifying leaf diseases on apple trees for major and minor classes, i.e., for multiple classification.
References
Agarwal M, Kaliyar RK, Singal G, Gupta SK (2019) FCNN-LDA: A faster convolution neural network model for leaf disease identification on apple’s leaf dataset. The International Conference on Information & Communication Technology and System, 246–251
Amara J, Bouaziz B, Algergawy A (2017) A deep learning-based approach for banana leaf diseases classification. Lecture Notes in Informatics 79–88
Arnal Barbedo JG (2014) An automatic method to detect and measure leaf disease symptoms using digital image processing. Plant Dis 98(12):1709–1716
Arnal Barbedo JG (2018) Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput Electron Agric 153:46–53
Ayaz H, Rodríguez-Esparza E, Ahmad M, Oliva D, Pérez-Cisneros M, Sarkar R (2021) Classification of apple disease based on non-linear deep features. Appl Sci 11(14):6422
Bhateja V, Gautam A, Tiwari A, Bao LN, Satapathy SC, Nhu NG, Le DN (2018) Haralick features-based classification of mammograms using SVM. In Information Systems Design and Intelligent Applications, Singapore, pp 787–795
Chaki J, Dey N, Moraru L, Shi F (2019) Fragmented plant leaf recognition: Bag-of-features, fuzzy-color and edge-texture histogram descriptors with multi-layer perceptron. Optik 181:639–650
Chao X, Sun G, Zhao H, Li M, He D (2020) Identification of apple tree leaf diseases based on deep learning models. Symmetry 12(7):1065
Dai P, Liang X, Wang Y, Gleason ML, Zhang R, Sun G (2019) High humidity and age-dependent fruit susceptibility promote development of trichothecium black spot on apple. Plant Dis 103(2):259–267
Guo X, Yin Y, Dong C, Yang G, Zhou G (2008) On the class imbalance problem. Int Conf Nat Comput 4:192–201
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, pp 770–778
Huber DM, Jones JB (2013) The role of magnesium in plant disease. Plant Soil 368(1):73–85
Husson O, Sarthou JP, Bousset L, Ratnadass A, Schmidt HP, Kempf P, Husson JKB, Tingry S, Aubertot JN, Deguine JP, Goebel FR, Lamichhane JR (2021) Soil and plant health in relation to dynamic sustainment of eh and ph homeostasis: A review. Plant Soil 466(1):391–447
Jadhav SB, Udupi VR, Patil SB (2020) Identification of plant diseases using convolutional neural networks. Int J Inf Technol 1–10
Mahlein AK (2016) Plant disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis 100(2):241–251
Militante SV, Gerardo BD, Dionisio NV (2019) Plant leaf detection and disease recognition using deep learning. IEEE Eurasia Conference on IOT, Communication and Engineering, pp 579–582
Moinina A, Lahlali R, MacLean D, Boulif M (2018) Farmers’ knowledge, perception and practices in apple pest management and climate change in the fes-meknes region, morocco. Horticulturae 4(4):42
Moinina A, Lahlali R, Boulif M (2019) Important pests, diseases and weather conditions affecting apple production in morocco: Current state and perspectives. Rev Marocaine Sci Agron Vet 7(1):71–87
Raj A, Srivastava A, Bhateja V (2011) Computer aided detection of brain tumor in magnetic resonance images. Int J Eng Technol 3(5):523
Raschka S (2018) Model evaluation, model selection, and algorithm selection in machine learning. arXiv preprint arXiv:1811.12808
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, Stefanovic D (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Comput Intell Neurosci 2016:3289801
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. IEEE Conference on Computer Vision and Pattern Recognition, pp 2818–2826
Tan M, Le Q (2019) Efficientnet: Rethinking model scaling for convolutional neural networks. International Conference on Machine Learning, pp 6105–6114
Thapa R, Zhang K, Snavely N, Belongie S, Khan A (2020) The plant pathology challenge 2020 data set to classify foliar disease of apples. Appl Plant Sci 8(9):e11390
Tharwat A, Gaber T, Awad YM, Dey N, Hassanien AE (2016) Plants identification using feature fusion technique and bagging classifier. The International Conference on Advanced Intelligent System, pp 461–471
Wöhner T, Emeriewen OF (2019) Apple blotch disease (marssonina coronaria (ellis & davis) davis)–review and research prospects. Eur J Plant Pathol 153(3):657–669
Yu HJ, Son CH, Lee DH (2020) Apple leaf disease identification through region-of-interest-aware deep convolutional neural network. J Imaging Sci Technol 64(2):20507–20511
Yun S, Han D, Oh SJ, Chun S, Choe J, Yoo Y (2019) Cutmix: Regularization strategy to train strong classifiers with localizable features. The IEEE/CVF International Conference on Computer Vision, pp 6023–6032
Zhang H, Cisse M, Fauphin YN, Lopez-Paz D (2017) Mixup: Beyond empirical risk minimization. arXiv preprint arXiv:1710.09412
Zhong Y, Zhao M (2020) Research on deep learning in apple leaf disease recognition. Comput Electron Agric 168:105146
Funding
Open access funding provided by Western Norway University Of Applied Sciences
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Muhammad Tariq.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yadav, A., Thakur, U., Saxena, R. et al. AFD-Net: Apple Foliar Disease multi classification using deep learning on plant pathology dataset. Plant Soil 477, 595–611 (2022). https://doi.org/10.1007/s11104-022-05407-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-022-05407-3