Abstract
Aims
Bio-organic fertilizer and different additives are widely applied to suppress soil-borne diseases. However, how different additives alter bulk soil microflora and thereby induce the healthy rhizospheric microflora remains unclear.
Methods
A 3-season field experiment containing four fertilization management programs (chemical fertilizer, organic fertilizer, amino acid organic fertilizer, and bio-organic fertilizer was conducted in a tomato production agroecosystem with high disease incidence to evaluate the induced efficacy. The bacterial and fungal microflora of bulk and rhizosphere soil influenced by different management programs were performed on the Illumina MiSeq platform. Principal coordinate analysis based on the Bray-Curtis distance metric was performed to compare the similarities and differences of the bacterial and fungal community compositions among all soil samples.
Results
Soil amended with organic fertilizer, amino acid organic fertilizer, and bio-organic fertilizer progressively and significantly suppressed tomato diseases in comparison with chemical fertilizer, and bio-organic fertilizer presented the best efficacy in all seasons. Interestingly, rhizospheric and bulk soil bacterial and fungal communities of the different fertilization management programs were separated from each other. Six bacterial and 10 fungal rhizospheric genera positively correlated with the same genera observed in bulk soil showing significant relationships with tomato disease incidence were observed, and functional strain SQR9 can be detected in the bulk and rhizosphere soils of bio-organic fertilizer treatments. Additionally, the redundancy analysis results showed the genera in treated chemical fertilizer bulk soil were dominated by Ralstonia and Fusarium, the abundances of which were highest and lowest in treated chemical fertilizer and bio-organic fertilizer rhizosphere, respectively.
Conclusions
This study provided insights into soil-borne disease suppression by bulk soil management and confirmed that alterations to the bulk soil microbiota by different organic additives played distinct roles in the formation of rhizospheric soil microflora for the suppression of disease.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Tomato (Solanum lycopersicum L.) is one of the most important vegetable crops cultivated worldwide (Singh and Siddiqui 2015). However, bacterial and Fusarium wilts caused by Ralstonia solanacearum (Smith) (Wei et al. 2011) and Fusarium oxysporum f. sp. lycopersici (Shanmugam et al. 2015), respectively, are two common diseases throughout tomato growing areas. More seriously, these two pathogens often infect tomato roots concurrently under field conditions, resulting in complex disease occurrence and severe yield loss every year (Liu et al. 2012).
The main cause for soil-borne disease incidence is an unbalanced soil microecosystem (Avis et al. 2008). Most soil-borne pathogens are adapted to survive in bulk soil, while the rhizosphere is the playground and infection court where pathogens establish a parasitic relationship with the plant (Raaijmakers et al. 2009). The pathogens need to grow saprophytically in the rhizosphere to reach their host or to achieve sufficient numbers on their host before they can infect host tissue (Berendsen et al. 2012). It has been reported that plants actively recruit beneficial soil microorganisms in their rhizosphere to counteract pathogen assault (Mendes et al. 2011) and that rhizospheric microbial communities are influenced by soil type, plant development, fertilizer management, and other environmental factors (Chaparro et al. 2014; Horwath et al. 1998). The outcome of these factors is the development of a rhizospheric microbial community that differs markedly from the source communities in bulk soil (Minz et al. 2013). Moreover, Bakker et al. (2015) has demonstrated that different soil amendments could manipulate different bulk microbial communities, which further shows that legacy effects of prior selections in microbiotas may continue to influence rhizospheric microbial community composition. Thus, there is a need to study the relationship between the legacy effects from bulk soil to the rhizosphere on plant disease.
Chemical soil fumigation, organic amendments, and biocontrol have been suggested as control methods for bacterial and Fusarium wilts (French 1994; Bonanomi et al. 2010). Chemical soil fumigation can reduce the abundance of pathogens quickly, but pathogens might rebound to a higher abundance again (Gamliel et al. 2000). An application of organic amendments could effectively control most soil-borne pathogens (Bailey and Lazarovits 2003), however, an increase in disease incidence was also observed in several other cases after the addition of organic amendments (Mazzola et al. 2001). Recently, more studies have been performed to examine the relationships between rhizospheric microorganisms and plant diseases, which recently became popular in plant pathology (Cai and Liao 2003). Our previous study revealed that application of bio-organic fertilizer could control tomato disease via the manipulation of the rhizospheric microbial communities (Wang et al. 2015). However, how bio-organic fertilizer alters bulk soil microflora and induces the development of rhizospheric microflora remains unclear.
In this study, a 3-season field experiment was conducted in a tomato production agroecosystem with high disease incidence to evaluate the suppression of tomato disease by the following four multi-level fertilization management programs: chemical fertilizer, organic fertilizer, amino acids organic fertilizer, and bio-organic fertilizer. Afterwards, bulk and rhizospheric soil microbiotas were surveyed using Illumina-MiSeq sequencing to seek out whether rhizospheric microbiotas affected by the different fertilizer management programs exhibited varying disease suppression and how these effects were induced by bulk microbiotas alterations.
Materials and methods
Field site and experiment description
The field experimental site was located at the Nanjing Institute of Vegetable Science, Nanjing, China (31°43′ N, 118°46′ E). This region has the tropical monsoon climate with an average annual temperature and precipitation of 15.4 °C and 1106 mm, respectively. Tomato has been continuously cropped in the field for several years and has suffered from high bacterial and Fusarium wilt disease presence. The oven-dried soil had a pH value of 7.08, and the contents of organic matter, total N, NH4-N, NO3-N, available P, and available K were 28.4 g/kg, 2.04 g/kg, 52.7 mg/kg, 544 mg/kg, 110 mg/kg, and 277 mg/kg, respectively.
A 3-season field experiment was performed from March 2014 to June 2015 and included the following four treatments: (1) CF treatment, soil amended with chemical fertilizer; (2) OF treatment, soil amended with organic fertilizer (chicken manure compost); (3) AOF treatment, soil amended with amino acids organic fertilizer; and (4) BF treatment, soil amended with bio-organic fertilizer. Each treatment had three randomized independent replications. The chicken manure compost was produced by Nantong Huinong Co. Ltd., Jiangsu, China by composting chicken manure at 30–70 °C for more than 20 days. The bio-organic fertilizer was produced by solid state fermentation according to Liu et al. (2016). In brief, pre-compost matured chicken manure added with 0.2 ml g−1 of compound liquid amino acids for 4 days (the pH of the mixture was 5.0–6.0), then antagonistic strain Bacillus amyloliquefaciens SQR9 (Cao et al. 2011) was inoculated into the mixture for a secondary fermentation with 4 days. After fermentation, the SQR9 cell concentration in the bio-organic fertilizer was greater than 1 × 108 CFU g−1 dry weight. The fertilizer without antagonistic bacteria inoculation and produced with the same procedure was regarded as amino acids organic fertilizer. All treatments were adjusted to the same amount of N (225 kg/ha), P (65 kg/ha) and K (150 kg/ha) for each season using mineral fertilizers as necessary. The N (urea) and K (K2SO4) fertilizers were applied as basal and supplementary fertilizer, while the P (calcium superphosphate), organic fertilizer, amino acids organic fertilizer and bio-organic fertilizer were only used as basal fertilizers. A detailed fertilization scheme is shown in Table S1.
Assay of tomato disease incidence and yield
Two months after the tomato plantlets were transplanted into the field, a bioassay for disease incidence including bacterial wilt disease, Fusarium wilt disease and the two-wilt disease complex was performed until the end of the experiment and was based on observations of typical wilt symptoms, including foliage chlorosis, necrosis and drooping of the leaves. Disease incidence was expressed as the percentage of diseased plants per total number of plants. For total tomato fruit yield of each plot, all mature tomato fruits were harvested and weighed. The disease incidence and fruit yield of each crop season (1st: spring season; 2nd: autumn season; 3rd: spring season) were analyzed in this study.
Soil sampling, DNA extraction, real-time PCR assay and soil chemical analysis
Soil sampling was performed in June 2015 during tomato harvesting. Briefly, 6 healthy tomato plants were randomly selected in each replicate plot, 3 of which were pooled to minimize the community variation. The bulk and rhizospheric soil samples were obtained according to Bakker et al. (2015). Thus, 6 bulk and rhizospheric soil samples were collected for each treatment. Finally, 5 bulk and rhizospheric soil samples for each treatment were randomly chosen for subsequent DNA extraction using the UltraClean Soil DNA Isolation Kits (MoBio Laboratories Inc., Carlsbad, USA) according to the manufacturer’s protocol. The quality and concentration of the DNA samples were determined using a spectrophotometer (NanoDrop 2000, USA). Total numbers of bacteria and fungi were quantified by Real-Time PCR with primers Eub338 and Eub 518 and primers ITS1f and 5.8 s, respectively, according to Liu et al. (2016). The copy numbers of SQR9 was also quantified by Real-Time PCR with primers SQR9F (5′-CATGAGATGGCGGGCTTT-3′) and SQR9R (5′-CGCATCCTCCCTGTCTTTG-3′) according to Qiu et al. (2014). Each sample was performed in three replicates, and the results were expressed as log (copies g−1) dry soil. All bulk soil chemical properties were determined according to Bao (2010).
Illumina MiSeq sequencing
The DNA of each soil sample served as the template for the amplification of the 16S rRNA gene and the ITS region. The V4 region of the bacterial 16S rRNA gene was amplified using primers 520F (5′-AYTGGGYDTAAAGNG-3′) (Ahmed et al. 2009) and 802R (5′-TACNVGGGTATCTAATCC-3′) (Ahmed et al. 2009), and ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) (Gardes and Bruns 1993) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) (White et al. 1990) were used for the ITS1 region of the fungal ITS gene. The barcodes of soil samples used to distinguish each sample in the pyrosequencing programs in this study are provided in Table S2. The programs of amplification and sequencing of the 16S and ITS genes were performed at Personal Biotechnology Co, Ltd. (Shanghai, China) on the Illumina MiSeq instrument (USA). All sequences were deposited in the NCBI Sequence Read Archive database with the accession number (SRP067366).
Bioinformatics analysis
Quality control and annotation of the raw sequences were performed according to Liu et al. (2016). A total of 36,473 16S rRNA and 41,614 ITS gene sequences for each sample were randomly selected for further bacterial and fungal microbial community analysis, respectively. To compare the similarities and differences of the bacterial and fungal community compositions among all soil samples, principal coordinate analysis (PCoA) based on the Bray-Curtis distance metric was performed using Mothur (Liu et al. 2016) and analysis of molecular variance (AMOVA) was performed to evaluate the significant differences in bacterial and fungal community structures among the four fertilizer treatments. AMOVA was used to compare the relative abundance of different groups according to the ordination base on OTU. In addition, Pearson’s correlation coefficient was used to evaluate the correlation between selected rhizospheric soil genera (relative abundance >0.1%) and tomato disease incidence. Afterwards, the relative abundances of genera that showed significant differences with disease incidence were further compared among the different fertilizer management programs in bulk and rhizospheric soil. Furthermore, Pearson’s correlation coefficient was used to evaluate the correlation between the relative abundances of these genera in bulk and rhizospheric soil samples. In order to examine the relationships among the bulk soil bacterial and fungal genera, samples and environmental variables, redundancy analysis (RDA) was carried out via the vegan package of R (version 3.2.2), bioEnv procedure was performed to select the best subset of environmental variables. In addition, the Mantel test was used to calculate the correlation between the selected soil characteristics and the selected microbial genera.
Statistical analysis
The differences among the different treatments were assessed using a one-way ANOVA analysis, and the calculated means were subjected to Duncan’s multiple range test at P < 0.05. All analyses were performed in SPSS v18.0 (SPSS Inc., USA).
Results
Effects of different fertilization management programs on disease incidence and tomato yield
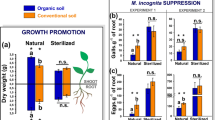
Disease incidence in the spring crop season in 2014 and 2015 was significantly higher (20–75%) than that in the autumn season (0–10%) in 2014 (Fig. 1a), and the disease incidence trends of all the seasons was similar [DI (CF) > DI (OF) > DI (AOF) > DI (BF)]. In the third season, the OF, AOF and BF treatments significantly (P < 0.05) reduced disease incidence to 30%, 16% and 6%, respectively. These results indicated that the bio-organic fertilizer application more effectively controlled the outbreak of wilt disease in tomato plants compared to the other treatments. In contrast to the disease incidences, tomato yields with the different fertilization treatments were significantly higher than CF in all crop seasons (Fig. 1b). For the third crop season, the application of organic fertilizer, amino acid organic fertilizer and bio-organic fertilizer significantly (P < 0.05) increased yield by 13%, 45% and 70%, respectively, compared to the CF treatment. These results indicated that the fertilization treatments (OF, AOF, and BF) progressively suppressed disease incidence and increased tomato crop yields compared to the CF treatment.
Total bacterial, fungal abundances and SQR9 copy numbers
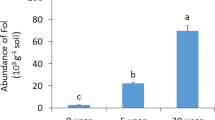
Compared to the CF treatment, the treatments of BF, AOF, and OF significantly (P < 0.05) increased bulk soil bacteria abundances (Fig. 2a). Similarly, rhizospheric bacteria abundances were also significantly (P < 0.05) enhanced. Moreover, no significant differences with fungi were observed in bulk or rhizospheric soils, regardless of fertilization management program (Fig. 2b). These results showed that the management programs that contained organic fertilizer (organic fertilizer, amino acid organic fertilizer and bio-organic fertilizer) had a positive effect on the abundance of bulk and rhizospheric bacteria rather than fungi compared to the CF treatment. The copy numbers (Log10 copies) of SQR9 in bulk and rhizosphere soils of BF treatment were 4.48 and 5.17 (Fig. 2c), respectively, while it cannot be detected in the other treatments of CF, OF and AOF.
Total bacterial (a) and fungal (b) microbial copies in bulk and rhizospheric soil with the different fertilization management programs. The copy numbers of SQR9 in the bulk and rhizosphere soil of BF treatment (c). Bars with different letters indicate significant differences as defined by Duncan’s test (P < 0.05). Fertilization management programs: chemical fertilizer (CF), organic fertilizer (OF), amino acids organic fertilizer (AOF) and bio-organic fertilizer (BF)
Sequencing results
As shown in Table S3, after basal quality control, a total of 2,171,613 16S rRNA and 3,548,914 ITS sequences were obtained for all soil samples. The number of high quality sequences per sample varied from 36,743 to 127,938 for bacteria and from 41,614 to 141,100 for fungi. Moreover, at the 97% similarity cut-off level, 7579 bacterial and 4517 fungal OTUs were obtained.
Microbial community composition
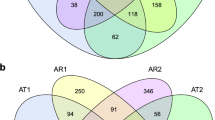
PCoA based on the Bray-Curtis distance metric revealed that the bulk soil bacterial (Fig. 3a) and fungal (Fig. 3b) communities significantly differed (P < 0.001) from those in the rhizosphere along the first component. Interestingly, the rhizospheric soil bacterial communities of all treatments were distinct (P < 0.05) from each other along the second component, showing the same separation tendency (P < 0.05) as the bulk soil. Similarly, the rhizospheric soil fungal communities for the different fertilization managements were all separated (P < 0.05) from each other along the second component with the same alteration trends (P < 0.05) for rhizospheric and bulk soil samples.
The bacterial (a) and fungal (b) microbial community compositions of the different treatments. Fertilization management programs: chemical fertilizer (CF), organic fertilizer (OF), amino acids organic fertilizer (AOF) and bio-organic fertilizer (BF); bulk soil amended with chemical fertilizer (BCF), organic fertilizer (BOF), amino acids organic fertilizer (BAOF) and bio-organic fertilizer (BBF); rhizosphere soil amended with chemical fertilizer (RCF), organic fertilizer (ROF), amino acids organic fertilizer (RAOF) and bio-organic fertilizer (RBF)
Genera abundance analysis
Clear positive correlations between disease incidence and the relative abundances of Bacillus (P < 0.05), Ralstonia (P < 0.05), Fimetariella (P < 0.01), Fusarium (P < 0.01), Gliomastix (P < 0.01), Guehomyces (P < 0.01), Humicola (P < 0.01), Penicillium (P < 0.01), and Trichoderma (P < 0.01), were observed. In contrast, negative correlations were observed for the genera Chitinophaga (P < 0.05), Enterobacter (P < 0.05), Pseudomonas (P < 0.05), Pseudoxanthomonas (P < 0.05), Debaryomyces (P < 0.01), Phialemonium, and Purpureocillium (P < 0.01) (Table 1).
Among these genera, the treatment of application of PGPR-containing organic fertilizer (BF) significantly (P < 0.05) increased the abundance of Chitinophaga, Pseudomonas, Debaryomyces, Phialemonium and Trichoderma and decreased the abundance of Humicola compared to the other treatments (CF, OF and AOF) in bulk soil (Fig. S1). Moreover, the lowest value of Fusarium was observed in the BF treatment, which was significantly (P < 0.05) lower than that in CF. Additionally, the AOF treatment showed the lowest relative abundance of Ralstonia and the highest relative abundance of Penicillium and Purpureocillium among all treatments.
Similar to bulk soil, the same variation trends of the relative abundances of Chitinophaga, Pseudomonas, Pseudoxanthomonas, Debaryomyces, Fusarium and Guehomyces were observed in the BF treated rhizosphere, and the highest and lowest values of the former four and latter two were observed. The AOF treatment significantly (P < 0.05) enriched the relative abundances of Debaryomyces and Purpureocillium compared to the CF and OF treatments. In addition, the organic fertilizer containing treatments (OF, AOF and BF) significantly reduced the relative abundance of Fusarium compared to CF and amended with bio-organic fertilizer showed significantly lower relative abundances of Ralstonia and Fusarium compared to other treatments.
Although the correlation coefficients of Ralstonia and Fusarium between bulk and rhizospheric soils were not significant (P > 0.05), the two genera including pathogens still showed a positive relationship (Fig. 4). Particularly, 8 genera of Chitinophaga, Pseudomonas, Pseudoxanthomonas, Debaryomyces, Guehomyces, Humicola, Phialemonium, and Purpureocillium in bulk soil had a significant (P < 0.01) and positive relationship with the corresponding genera in rhizosphere soil, and no genus showed a significantly (P < 0.05) negative relationship between the bulk and rhizospheric soils, indicating that the bulk microbiota is crucial and critical to the development of the rhizospheric microflora and suppressing tomato disease.
Effects of soil chemical properties on bacterial or fungal taxa
The three crop seasons with the different fertilization management programs changed the soil chemical properties (Table 2). The fertilization management programs containing organic fertilizer (OF, AOF and BF) significantly (P < 0.05) increased soil OM, TP and AP contents compared to the CF treatment. In addition, fertilizer management programs containing amino acids (AOF and BF) significantly (P < 0.05) enhanced soil EC and decreased soil TK and NH4-N concentrations compared to the fertilizer management programs that did not contain amino acids (CF and OF). Moreover, the BF treatment showed significantly (P < 0.05) higher NO3-N, TN, and AK contents and a lower pH value compared to the other treatments.
The Mantel test based on the selected soil chemical properties and the abundances of the analyzed microbial genera revealed that the selected soil chemical properties were significantly correlated with variations in the selected bacterial and fungal genera (r = 0.85, P < 0.001). The redundancy analysis performed to examine the relationship between the analyzed genera from bulk soil and soil chemical properties showed that the two components could explained 49.07% of the total variation (Fig. 5). The second component (RDA2), which explained 16.36% of the variation, separated BF from the other treatments. As shown in Fig. 5, the microbial genera in the BF soil samples were dominated by Trichoderma, Pseudomonas, Chitinophaga and were related to TN, EC and OM contents, while the genera in the CF soil samples were dominated by Ralstonia and Fusarium. Moreover, the microbial community in the AOF were dominated by Penicillium, Purpureocillium and Fimetariella; and in OF soil samples were dominated by Bacillus, Humicola and Fimetariella.
Redundancy analysis of the relationship between the analyzed bulk genera, samples and environmental variables. Fertilization management programs: chemical fertilizer (CF), organic fertilizer (OF), amino-acids organic fertilizer (AOF) and bio-organic fertilizer (BF). Bulk soil amended with chemical fertilizer (BCF), organic fertilizer (BOF), amino acids organic fertilizer (BAOF) and bio-organic fertilizer (BBF)
Discussion
Applications of organic fertilizer, amino acid organic fertilizer, and bio-organic fertilizer progressively and significantly suppressed plant disease and improved fruit yield compared to the chemical fertilizer. These results were in agreement with previous studies suggested that organic amendments could be used to control diseases caused by soil-borne pathogens (Hoitink and Fahy 1986; Szczech 1999). More importantly, the most efficacious disease suppression was observed in the BIO treatment, which is in accordance with previous reports where bio-organic fertilizer acted as both an organic fertilizer and a biocontrol agent for different soil-borne diseases in tomato (Wang et al. 2015), banana (Shen et al. 2013), and watermelon (Ling et al. 2014) when compared to organic amendments and biocontrol agents applied alone.
The fertilizer management programs (OF, AOF and BF) all significantly increased bacterial abundances when compared with CF and was similar to previous studies where higher populations of bacteria in organic treated soils were observed compared with that from a chemical fertilizer treatment (Witter et al. 1993). Interestingly, bacterial abundances in rhizospheric soils of these treatments were also higher compared to CF, indicating that bulk soil management induced microbial abundance variation in the rhizosphere. Functional strain SQR9 can be detected in the bulk and rhizosphere soils of BF treatments, suggesting that SQR9 could survive in the tomato bulk soil and further colonize in the rhizosphere soil. This result was in accordance with the other report as SQR9 could effectively colonize in the rhizosphere soil and suppress the cucumber plant disease (Qiu et al. 2014).
PCoA results revealed that the bacterial and fungal communities in the bulk soils of the different treatments were well differentiated from the rhizospheric communities along the first component, which were mainly due to the ability of root exudates to influence the composition of the rhizospheric microbial communities (Chaparro et al. 2014). Interestingly, in both bulk and rhizospheric soils, bacterial and fungal microbial communities of the different fertilization management programs were well separated from each other along the second component. In accordance with our results, Bonanomi et al. (2010) also reported that an application of organic amendments significantly shifted the soil microbial community. Moreover, amino acids are excellent C and N sources for microbes (Ge et al. 2009), and may induced microbial communities variation. Lots of researches have confirmed that the PGPR-containing bio-organic fertilizers had a considerable effect on shaping the microbial community (Wang et al. 2015; Shen et al. 2013; Ling et al. 2014). The same alteration trends for the bacterial and fungal communities for rhizospheric and bulk soil samples were observed. Ridder-Duine et al. (2005) also demonstrated that the rhizosphere microbial composition of the wild plant C. arenaria was largely dependent on the microbial composition of the bulk soil. Hence, we can deduce that the different fertilization management programs altered the microbial community in bulk soil first, inducing their specific rhizospheric microbiotas.
The rhizospheric genera Ralstonia, Fusarium, Bacillus, Fimetariella, Gliomastix, Guehomyces, Humicola, Penicillium, and Trichoderma were positively and significantly (P < 0.05) correlated with tomato disease incidence. In contrast, a significantly (P < 0.05) negative relationship was observed for genera Chitinophaga, Enterobacter, Pseudomonas, Pseudoxanthomonas, Debaryomyces, Phialemonium, and Purpureocillium. Because all the tested plants in the different treatments were healthy, the healthy plants in high disease incidence treatments contained more relative abundances of the rhizospheric genera Ralstonia and Fusarium, increasing the risk of plant illness (Wei et al. 2011; Shanmugam et al. 2015). Moreover, studies have confirmed that the genera Bacillus (Singh and Siddiqui 2015), Penicillium (Sabuquillo et al. 2006), and Trichoderma (Blaya et al. 2013) act as biocontrol agents could induce a resistance capacity of plants that provides protection against of the microbial pathogens (Ent et al. 2009). Pseudomonas was reported to suppress plant disease in several studies (Dowling and O'Gara 1994; Singh and Siddiqui 2015). Enterobacter has been reported for bioactivity against Fusarium (Al-Mughrabi 2010). Pseudoxanthomonas is known as a biocontrol agent against Xanthomonas (Al-Saleh 2014). Debaryomyces has been reported as a biocontrol agent against Penicillium expansum (Singh et al. 2011), and Purpureocillium has been found exhibiting bio-control of Meloidogyne incognita (Singh et al. 2013). However, the role played by these three genera for suppressing Ralstonia and Fusarium are still unclear. Xue et al. (2015) reported that the relative abundance of Bacillus was negatively correlated with banana disease incidence, while in this study, the relative abundance of Bacillus was positively correlated with pathogen abundance and disease incidence as well as Trichoderma. Previous study observed when plant infected by pathogens, plant roots would secrete more citric acid and fumaric acid to stimulate the chemotaxis response of plant growth promoting rhizobacteria (Liu et al. 2014). This may be the reason why more Bacillus and Trichoderma occurred in the higher plant disease incidence rhizosphere soil. Furthermore, of the 16 rhizospheric genera, 8 showed a significant (P < 0.01) positive relationship to the abundances of the same genera in bulk soil, while none of them showed a significantly (P < 0.05) negative relationship. Similar dynamics were observed across the communities, such as the relative abundance of Actinobacteria, Firmicutes, Proteobacteria, Bacteroidetes and Acidobacteria, as the rhizosphere developed from bulk soil communities (Bakker et al. 2015). Therefore, our results suggested that the bulk microbial community played the crucial and critical role in manipulating the rhizospheric microflora and suppressing plant disease.
The fertilizer management programs containing organic fertilizer (OF, AOF and BF) significantly (P < 0.05) increased soil OM compared to CF, which were consistent with a previous study (Haynes and Naidu 1998). The fertilizer management programs containing amino acids (AOF and BF) significantly (P < 0.05) enhanced soil EC and decreased NH4-N compared to the fertilizer management programs containing no amino acids (CF and OF). Amino acids can form complexes with cations via carboxylate (−COO) and amine (−NH2) groups (Dalir and Khoshgoftarmanesh 2014) and might be the reason for the enhancement of soil EC. Yadessa et al. (2010) also found that there were significantly (P < 0.01) negative correlations between tomato bacterial wilt disease incidence and EC. The higher soil NH4-N content in the fertilizer treatments with no amino acids may be due to the application of the chemical nitrogen (urea breaks down to NH4-N) fertilizer (Shen et al. 2013). Moreover, the BF treatment showed significantly (P < 0.05) higher soil NO3-N content than the CF, OF and AOF treatments. Huber and Watson (1974) reviewed that tomato root disease caused by Fusarium could be reduced by decreased NO3-N and increased NH4-N contents. In contrast, it was reported that bacterial wilt of tomato increased as NO3-N increased and was reduced as NH4-N was reduced (Yadessa et al. 2010). Although the processes of disease suppression were different, the possible key factor of disease suppression was to decrease the rhizospheric population of pathogens (Nel et al. 2007). Additionally, the genera in CF bulk soil were dominated by Ralstonia and Fusarium. While, this phenomena was not observed in OF, AOF and BF treatments. Those results also suggested that soil amended with organic fertilizers have a high efficacy for suppressing the diseases. Strikingly, the application of bio-organic fertilizer showed the highest efficacy.
Conclusion
In the present study, the application of different additives in a three-season field experiment showed fertilization management programs (organic fertilizer, amino acid organic fertilizer and bio-organic fertilizer) progressively and significantly decreased soil-borne diseases and enhanced fruit yields of tomato compared to the CF treatment. Bulk soil microbial compositions in agroecosystems were significantly affected by the different fertilizer management programs, and the altered bulk soil microbial communities played a crucial role in manipulating rhizospheric soil microflora. Compared to the CF treatment, OF, AOF and BF had a high efficacy for suppressing the tomato disease, while BF possessed the highest efficacy. This study provides insights into the soil-borne disease suppression by bulk soil management to induce healthy rhizospheric soil microflora.
References
Ahmed N, Claesson MJ, O’Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O’Toole PW (2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669
Al-Mughrabi KI (2010) Biological control of Fusarium dry rot and other potato tuber diseases using Pseudomonas fluorescens, and Enterobacter cloacae. Biol Control 53:280–284
Al-Saleh MA (2014) Evaluation of saudi fluorescent Pseudomonads isolates as a biocontrol agent against citrus canker disease caused by Xanthomonas citri subsp citri a. Egypt Acad J Biolog Sci 6(2):1–7
Avis TJ, Gravel V, Antoun H, Tweddell R (2008) Multifaceted beneficial effects of rhizosphere microorganismson plant health and productivity. Soil Biol Biochem 40:1733–1740
Bailey KL, Lazarovits G (2003) Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res 72:169–180
Bakker MG, Chaparro JM, Manter DK, Vivanco JM (2015) Impacts of bulk soil microbial community structure on rhizosphere microbiomes of zea mays. Plant Soil 392:115–126
Bao S (2010) Soil and agro-chemical analytical methods. China Agriculture Press, Beijing
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Blaya J, López-Mondéjar R, Lloret E, Pascual JA, Ros M (2013) Changes induced by Trichoderma harzianum in suppressive compost controlling fusarium wilt. Pestic Biochem Physiol 107:112–119
Bonanomi G, Antignani V, Capodilupo M, Scala F (2010) Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biol Biochem 42:136–144
Cai YF, Liao ZW (2003) Effect of fertilization on the control of tomato bacterial wilt and soil health restoration using FAME analysis. Sci Agric Sin 8:010
Cao Y, Zhang ZH, Ling N, Yuan YJ, Zheng XY, Shen B, Shen QR (2011) Bacillus subtilis SQR9 can control fusarium wilt in cucumber by colonizing plant roots. Biol Fertil Soils 47:495–506
Chaparro JM, Badri DV, Vivanco JM (2014) Rhizospheric microbiome assemblage is affected by plant development. ISME J 8:790–803
Dalir N, Khoshgoftarmanesh AH (2014) Symplastic and apoplastic uptake and root to shoot translocation of nickel in wheat as affected by exogenous amino acids. J Plant Physiol 171:531–536
Dowling DN, O'Gara F (1994) Metabolites of pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol 12:133–141
Ent SVD, Wees SCMV, Pieterse CMJ (2009) Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70:1581–1588
French ER (1994) Integrated control of bacterial wilt of potato. CIP Circular 20:8–11
Gamliel A, Austerweil M, Kritzman G (2000) Non-chemical approach to soilborne pest management–organic amendments. Crop Prot 19:847–853
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Ge T, Song SW, Roberts P, Jones DL, Huang DF, Iwasaki K (2009) Amino acids as a nitrogen source for tomato seedlings: the use of dual-labeled (13 C, 15 N) glycine to test for direct uptake by tomato seedlings. Environ Exp Bot 66:357–361
Haynes RJ, Naidu R (1998) Influence of lime, fertilizer and manure applications on soil organic matter content and soil physical conditions: a review. Nutr Cycl Agroecosyst 51:123–137
Hoitink HAJ, Fahy PC (1986) Basis for the control of soil borne plant pathogens with composts. Annu Rev Phytopathol 24:93–114
Horwath WR, Elliott LF, Lynch JM (1998) Influence of soil quality on the function of inhibitory rhizobacteria. Lett Appl Microbiol 26:87–92
Huber DM, Watson RD (1974) Nitrogen form and plant disease. Annu Rev Phytopathol 12:139–165
Ling N, Deng KY, Song Y, YC W, Zhao J, Raza W, Shen QR (2014) Variation of rhizospheric bacterial community in watermelon continuous mono-cropping soil by long-term application of a novel bioorganic fertilizer. Microbiol Res 169:570–578
Liu YZ, Chen ZY, Liang XJ, Zhu J (2012) Screening, evaluation and identification of antagonistic bacteria against Fusarium oxysporum f. sp. lycopersici and Ralstonia solanacearum. Chinese Journal of Biological Control 28:101–108
Liu YP, Zhang N, Qiu MH, Feng HC, Vivanco JM, Shen QR, Zhang RF (2014) Enhanced rhizosphere colonization of beneficial bacillus amyloliquefaciens sqr9 by pathogen infection. FEMS Microbiol Lett 353:49–56
Liu HJ, Chen DD, Zhang RF, Hang XN, Li R (2016) Amino acids hydrolyzed from animal carcasses are a good additive for the production of bio-organic fertilizer. Front Microbiol 7:e6669
Mazzola M, Granatstein DM, Elfving DC, Mullinix K (2001) Suppression of specific apple root pathogens by Brassica napus seed meal amendment regardless of glucosinolate content. Phytopathology 91:673–679
Mendes R, Kruijt M, de Bruijn I, Dekkers, van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM (2011) Deciphering the rhizospheric microbiome for disease-suppressive bacteria. Science 332:1097–1100
Minz D, Ofek M, Hadar Y (2013) Plant rhizosphere microbial communities. Springer, Berlin
Nel B, Steinberg C, Labuschagne N, Viljoen A (2007) Evaluation of fungicides and sterilants for potential application in the management of fusarium wilt of banana. Crop Prot 26:697–705
Qiu MH, Li SQ, Zhou X, Cui XS, Vivanco JM, Zhang N, Shen QR, Zhang RF (2014) De-coupling of root–microbiome associations followed by antagonist inoculation improves rhizosphere soil suppressiveness. Biol Fertil Soils 50:217–224
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361
Ridder-Duine ASD, Kowalchuk GA, Gunnewiek PJAK, Smant W, Veen JAV, Boer WD (2005) Rhizosphere bacterial community composition in natural stands of Carex arenaria, (sand sedge) is determined by bulk soil community composition. Soil Biol Biochem 37:349–357
Sabuquillo P, Ade C, Melgarejo P (2006) Dispersal improvement of a powder formulation of Penicillium oxalicum, a biocontrol agent of tomato wilt. Plant Dis 89:1317–1323
Shanmugam V, Chugh P, Sharma P (2015) Cold-tolerant Trichoderma species for the management of fusarium wilt of tomato plants. Ann Microbiol 65:543–551
Shen ZZ, Zhong ST, Wang YG, Wang BB, Mei XL, Li R, Shen QR (2013) Induced soil microbial suppression of banana fusarium wilt disease using compost and biofertilizers to improve yield and quality. Eur J Soil Biol 57:1–8
Singh N, Siddiqui ZA (2015) Effects of Bacillus subtilis, Pseudomonas fluorescens and Aspergillus awamori on the wilt-leaf spot disease complex of tomato. Phytoparasitica 43:61–75
Singh D, Sharma RR, Samuel DVK, Pal RK (2011) Enhancing the bio-efficacy of Debaryomyces hansenii with sodium salts for reducing the blue mould rot in apples. Indian. Phytopathology 4:478–483
Singh S, Pandey RK, Goswami BK (2013) Bio-control activity of Purpureocillium lilacinum strains in managing root-knot disease of tomato caused by Meloidogyne incognita. Biomed Sci Technol 23:1469–1489
Szczech MM (1999) Suppressiveness of vermicompost against fusarium wilt of tomato. J Phytopathol 147:155–161
Wang LF, Lu XP, Yuan HY, Wang B, Shen QR (2015) Application of bio-organic fertilizer to control tomato fusarium wilting by manipulating soil microbial communities and development. Commun Soil Sci Plant 46:2311–2322
Wei Z, Yang XM, Yin SX, Shen QR, Ran W, YC X (2011) Efficacy of Bacillus-fortified organic fertiliser in controlling bacterial wilt of tomato in the field. Appl Soil Ecol 48:152–159
White TJ, Bruns TD, Lee SB, Taylor JW, Innis MA, Gelfand DH (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for Phylogenetics. PCR Protocols:315–322
Witter E, Mårtensson AM, Garcia FV (1993) Size of the soil microbial biomass in a long-term field experiment as affected by different n-fertilizers and organic manures. Soil Biol Biochem 25:659–669
Xue C, Penton CR, Shen ZZ, Zhang RF, Huang QW, Li R, Ruan YZ, Shen QR (2015) Manipulating the banana rhizosphere microbiome for biological control of panama disease. Sci Rep 5:11124
Yadessa GB, Van Bruggen AHC, Ocho FL (2010) Effects of different soil amendments on bacterial wilt caused by Ralstonia solanacearum and on the yield of tomato. J Plant Pathol 92:439–450
Acknowledgements
This research was supported by the National Key Basic Research Program of China (2015CB150506), the National key research and development program (2016YFD0800605, 2016YFE0101100 and 2016YFD0200106), the National Natural Science Foundation of China (41571242), Jiangsu Science and Technology Program (BE2014398, BY2016077-05), the Natural Science Foundation of Jiangsu Province, China (BK20160710 and BK20150059), the Priority Academic Program Development of the Jiangsu Higher Education Institutions (PAPD), 111 project (B12009), and Jiangsu Key Laboratory for Solid Organic Waste Utilization (BM2011013).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Birgit Mitter
Electronic supplementary material
Figure S1
Bulk (A) and rhizospheric (B) relative abundance of the analyzed genera. Fertilization management programs: chemical fertilizer (CF), organic fertilizer (OF), amino acids organic fertilizer (AOF) and bio-organic fertilizer (BF). Bulk soil amended with chemical fertilizer (BCF), organic fertilizer (BOF), amino acids organic fertilizer (BAOF) and bio-organic fertilizer (BBF). Rhizosphere soil amended with chemical fertilizer (RCF), organic fertilizer (ROF), amino acids organic fertilizer (RAOF) and bio-organic fertilizer (RBF). (GIF 22 kb)
Table S1
(DOC 35 kb)
Table S2
(DOC 59 kb)
Table S3
(DOC 55 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liu, H., Xiong, W., Zhang, R. et al. Continuous application of different organic additives can suppress tomato disease by inducing the healthy rhizospheric microbiota through alterations to the bulk soil microflora. Plant Soil 423, 229–240 (2018). https://doi.org/10.1007/s11104-017-3504-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3504-6