Abstract
Background and Aims
Root branching leads to morphological and functional heterogeneity of fine roots. However, the structure of soil microbiota associated with root branching orders has never been investigated. Deep insights into rhizosphere microbial community could provide a better understanding of the plant-microbe relationship.
Methods
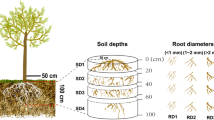
Fine roots of poplar (Populus × euramericana (Dode) Guinier.) were sampled and sorted into three groups according to their branching orders. Scanning electron microscopy (SEM) was used to observe the surface features of different orders of fine roots. Illumina MiSeq was employed to analyze the bacterial community structure of soil compartments from different root orders (i.e., R1, R2, and R3) and bulk soil compartment (NR).
Results
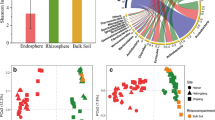
SEM showed that the first-order root with smaller diameter had dense coverage of vigorous root hairs, whereas higher order roots with larger diameter had sloughed-off cortical tissues on the rhizoplane. The diversity of bacterial communities was higher in the R1 and R2 compartment than in R3 or NR. There were 80 genera with a relative abundance above 0.05% in soils. Ternary plot revealed that bacterial genera were significantly enriched in R1 than in R2 or R3. Redundant analysis (RDA) showed that 12 dominant bacterial genera with a relative abundance over 1% were significantly correlated with P and NH4 +-N content of soils.
Conclusions
Root orders could influence the inhabitation of bacterial communities. The core groups of bacterial communities inhabiting the rhizosphere were correlated with soil nutrients. The root order-dependent interactions of plant-microbe provided a new model about the association between roots and soils.






Similar content being viewed by others
References
Badri DV, Vivanco JM (2009) Regulation and function of root exudates. Plant Cell Environ 32:666–681
Bagniewska-Zadworna A, Stelmasik A, Minicka J (2014) From birth to death – Populus trichocarpa fibrous roots functional anatomy. Biol Plant 58:551–560
Barlow PW (1975) The root cap. In: Torrey JG, Clarkson DT (eds) The development and function of roots (Third Cabot Symposium). Academic, London, pp 21–54
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Berg G, Zachow C, Lottmann J, Götz M, Costa R, Smalla K (2005) Impact of plant species and site on rhizosphere–associated fungi antagonistic to Verticillium dahliae Kleb. Appl Environ Microbiol 71:4203–4213
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality–filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
Braak T, Šmilauer PN (2002) Canoco reference manual and CanoDraw for Windows: user’ guide: software for Canonical community ordination (version 4.5). Microcomputer Power, Ithaca
Bulgarelli D, Rot M, Schlaeppi K, Themaat EVLV, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root–inhabiting bacterial microbiota. Nature 488:91–95
Caldwell MM, Dawson TE, Richards JH (1998) Hydraulic lift: consequences of water efflux from the roots of plants. Oecologia 113:151–161
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N et al (2010) QIIME allows analysis of high–throughput community sequencing data. Nat Methods 7:335–336
Cheng W, Parton WJ, Gonzalez-Meler MA, Phillips R, Asao S, McNickle G, Brzostek E, Jastrow JD (2014) Synthesis and modeling perspective of rhizosphere priming. New Phytol 201:31–44
Desantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera–checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Dijkstra FA, Carrillo Y, Pendall E, Morgan JA (2013) Rhizosphere priming: a nutrient perspective. Front Microbiol 4:1–8
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Eisenhauer N, Scheu S, Jousset A (2012) Bacterial diversity stabilizes community productivity. PLoS One 7:e34517
Fan PP, Guo DL (2010) Slow decomposition of lower order roots: a key mechanism of root carbon and nutrient retention in the soil. Oecologia 163:509–515
Farrar J, Hawes M, Jones D, Lindow S (2003) How roots control the flux of carbon to the rhizosphere. Ecology 84:827–837
Gordon WS, Jackson RB (2000) Nutrient concentrations in fine roots. Ecology 81:275–280
Gottel NR, Castro HF, Kerley M, Yang Z, Pelletier DA, Podar M, Karpinets T, Uberbacher E, Tuskan GA, Vilgalys R, Doktycz MJ, Schadt CW (2011) Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microbiol 77:5934–5944
Grayston SJ, Griffith GS, Mawdslcy JL, Campbell CD, Bardgett RD (2001) Accounting for variability in soil microbial communities of temperate upland grassland ecosystem. Soil Biol Biochem 33:533–551
Grayston SJ, Wang S, Campbell CD, Edwards AC (1998) Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem 30:369–378
Guo DL, Li H, Mitchell RJ, Han WX, Hendricks JJ, Fahey TJ, Hendrick RL (2008a) Fine root heterogeneity by branch order: Exploring the discrepancy in root turnover estimates between minirhizotron and carbon isotopic methods. New Phytol 177:443–456
Guo DL, Xia MX, Wei X, Chang WJ, Liu Y, Wang ZQ (2008b) Anatomical traits associated with absorption and mycorrhical colonization are linked to root branch order in twenty–three Chinese temperate tree species. New Phytol 180:673–683
Haichar FZ, Santaella C, Heulin T, Achouak W (2014) Root exudates mediated interactions belowground. Soil Biol Biochem 77:69–80
Hajek P, Hertel D, Leuschner C (2014) Root order– and root age–dependent response of two poplar species to belowground competition. Plant Soil 377:337–355
Hartmann A, Schmid M, Tuinen DV, Berg G (2009) Plant–driven selection of microbes. Plant Soil 321:235–257
Hishi T, Takeda H (2005) Dynamics of heterorhizic root systems: Protoxylem groups within the fine–root system of Chamaecyparis obtuse. New Phytol 167:509–521
Iversen CM (2014) Using root form to improve our understanding of root function. New Phytol 203:707–709
Jackson RB, Mooney HA, Schulze ED (1997) A global budget for fine root biomass, surface area, and nutrient contents. Proc Natl Acad Sci U S A 94:7362–7366
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3:442–453
Kong DL, Ma CG, Zhang Q, Li L, Chen XY, Zeng H, Guo DL (2014) Leading dimensions in absorptive root trait variation across 96 subtropical forest species. New Phytol 203:863–872
Kuzyakov Y (2010) Priming effects: interactions between living and dead organic matter. Soil Biol Biochem 42:1363–1371
Kuzyakov Y, Xu X (2013) Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytol 198:656–669
Li XZ, Rui JP, Mao YJ, Yannarell A, Mackie R (2014) Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol Biochem 68:392–401
Ling N, Yang S, Raza W, Huang Q, Guo S, Shen Q (2015) The response of root–associated bacterial community to the grafting of watermelon. Plant Soil 391:352–264
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
McCormack ML, Dickie IA, Eissensat DM, Fahey TJ, Fernandez CW, Guo DL, Helmisaari HS, Hobbie EA, Iversen CM, Jackson RB, Leppälammi-Kujansuu J, Norby RJ, Phillips RP, Pregitzer KS, Pritchard SG, Rewald R, Zadworny M (2015) Redefining fine roots improves understanding of below–ground contributions to terrestrial biosphere processes. New Phytol 207:505–518
Meng H, Zhang Y, Zhao LL, Zhao WJ, He C, Honaker CF, Zhai ZX, Sun ZK, Siegel PB (2014) Body weight selection affects quantitative genetic correlated responses in gut microbiota. PLoS One 9:e89862
Michael AQ, Miriam S, Paul C, Thomas DO, Simon RH, Thomas RC, Anna BE, Harold PS, Yong G (2012) A tale of three next generation sequencing platforms: comparison of ion torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341
Osmont KS, Sibout R, Hardtke CS (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58:93–113
Pan F, Xu A, Xia D, Yu Y, Chen G, Meyer M, Zhao D, Huang CH, Wu Q, Fu J (2015) Effects of octahedral molecular sieve on treatment performance, microbial metabolism, and microbial community in expanded granular sludge bed reactor. Water Res 87:127–136
Paterson E, Gebbing T, Abel C, Sim A, Telfer G (2007) Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol 173:600–610
Pires AC, Cleary DR, Almeida A, Cunha Â, Dealtry S, Mendonça-Hagler LS, Smalla K, Gomesa NM (2012) Denaturing gradient gel electrophoresis and barcoded pyrosequencing reveal unprecedented archaeal diversity in mangrove sediment and rhizosphere samples. Appl Environ Microbiol 78:5520–5528
Pregitzer KS, Kubiske ME, Yu CK, Hendrick RL (1997) Relationships among root branch order, carbon, and nitrogen in four temperate species. Oecologia 111:302–308
Pregitzer KS, De Forest JL, Burton AJ (2002) Fine root architecture of nine north American trees. Ecol Monogr 72:293–309
Rutvisuttinunt W, Chinnawirotpisan P, Simasathien S, Shrestha SK, Yoon IK, Klungthong C, Fernandez S (2013) Simultaneous and complete genome sequencing of influenza A and B with high coverage by Illumina MiSeq platform. J Virol Methods 193:394–404
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl GW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open–source, platform–independent, community–supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Shakya M, Gottel N, Castro H, Yang ZK, Gunter L, Labbé J, Muchero W, Bonito G, Vilgalys R, Tuskan G, Podar M, Schadt CW (2013) A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS One 8:e76382
Timonen S, Hurek T (2006) Characterization of culturable bacterial populations associating with Pinus sylvestris– Suillus bovinus mycorrhizospheres. Can J Microbiol 52:769–778
Uroz S, Buee M, Murat C, Frey-Klett P, Martin F (2010) Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2:281–288
Vogt KA, Vogt DJ, Bloomfield J (1998) Analysis of some direct and indirect methods for estimating root biomass and production of forests at an ecosystem level. Plant Soil 200:71–89
Wang YP, Li CR, Wang QK, Wang HT, Duan BL, Zhang GC (2016) Environmental behaviors of phenolic acids dominated their rhizodeposition in boreal poplar plantation forest soils. J Soils Sediments 16:1858–1870
Watteau F, Villemin G, Ghanbaja J, Genet P, Pargney JC (2002) In situ ageing of fine beech roots (Fagus sylvatica) assessed by transmission electron microscopy and electron energy loss spectroscopy: description of microsites and evolution of polyphenolic substances. Biol Cell 94:55–63
Wells CE, Eissenstat DM (2002) Beyond the roots of young seedlings: the influence of age and order on fine root physiology. J Plant Growth Regul 21:324–334
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer Press, London
Williams S, Foster P, Littlewood D (2014) The complete mitochondrial genome of a turbinid vetigastropod from MiSeq Illumina sequencing of genomic DNA and steps towards a resolved gastropod phylogeny. Gene 533:38–47
Xia MX, Guo DL, Pregitzer KS (2010) Ephemeral root modules in Fraxinus mandshurica. New Phytol 188:1065–1074
Xu T, Wang HT, Zhu WR, Wang YP, Li CR, Jiang YZ (2015) Morphological and anatomical traits of poplar fine roots in successive rotation plantations. Sci Silvae Sin 51:119–126
Yoon-Seong J, Sang-Cheol P, Jeongmin L, Jongsik C, Bong-Soo K (2015) Improved pipeline for reducing erroneous identification by 16S rRNA sequences using the Illumina MiSeq platform. J Microbiol 53:60–69
Zadworny M, McCormack ML, Rawlik K, Jagodzinski AM (2015) Seasonal variation in chemistry, but not morphology, in roots of Quercus robur growing in different soil types. Tree Physiol 35:644–652
Zhao J, Ni T, Li Y, Xiong W, Ran W, Shen B, Shen Q, Zhang R (2014) Responses of bacterial communities in arable soils in a rice–wheat cropping system to different fertilizer regimes and sampling times. PLoS One 9:e85301
Zhu B, Gutknecht JM, Herman DJ, Keck DC, Firestone MK, Cheng WX (2014) Rhizosphere priming effects on soil carbon and nitrogen mineralization. Soil Biol Biochem 76:183–192
Acknowledgments
This work was funded by the National Basic Research Program of China (2012CB416904) and the National Natural Science Foundation of China (31570618). Additionally, this study was supported by the funds of the Shandong Double Tops Program (Grant No. SYL2017XTTD03). We thank the anonymous reviewers for the helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Stéphane Compant
Electronic supplementary material
ESM 1
(DOC 12988 kb)
Rights and permissions
About this article
Cite this article
Wang, Q., Wang, N., Wang, Y. et al. Differences in root-associated bacterial communities among fine root branching orders of poplar (Populus × euramericana (Dode) Guinier.). Plant Soil 421, 123–135 (2017). https://doi.org/10.1007/s11104-017-3449-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3449-9